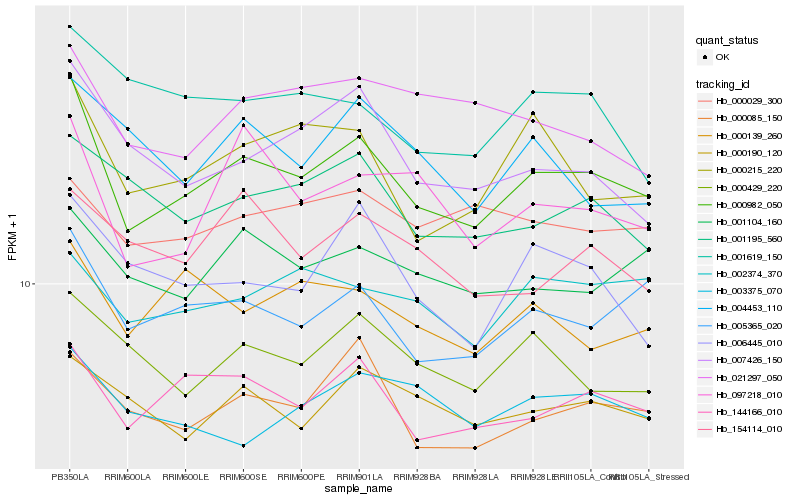
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000982_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643124 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005365_020 |
0.079965568 |
- |
- |
hypothetical protein JCGZ_15105 [Jatropha curcas] |
| 3 |
Hb_144166_010 |
0.0824533291 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 4 |
Hb_000215_220 |
0.0920124868 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000085_150 |
0.0931276871 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 6 |
Hb_007426_150 |
0.0959711065 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 7 |
Hb_001104_160 |
0.0991174882 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 8 |
Hb_097218_010 |
0.0993879217 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000429_220 |
0.0993992231 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001195_560 |
0.1012349895 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000190_120 |
0.1028657118 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 12 |
Hb_000139_260 |
0.1033227832 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 13 |
Hb_006445_010 |
0.1056403535 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 14 |
Hb_002374_370 |
0.106152653 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_003375_070 |
0.1095020544 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 16 |
Hb_021297_050 |
0.1095945011 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 17 |
Hb_154114_010 |
0.1105990518 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 18 |
Hb_004453_110 |
0.1109230526 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 19 |
Hb_001619_150 |
0.1111465941 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000029_300 |
0.1112179949 |
- |
- |
hypothetical protein JCGZ_10048 [Jatropha curcas] |