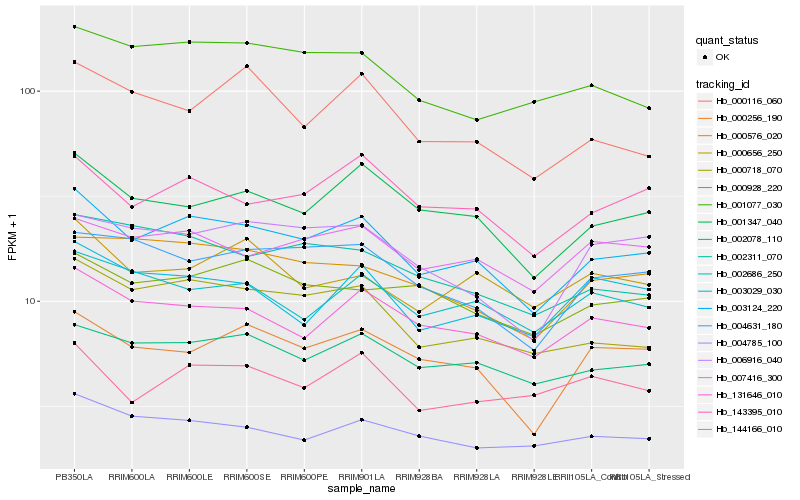
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_220 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 2 |
Hb_000718_070 |
0.0660164667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001347_040 |
0.0660555547 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 4 |
Hb_002078_110 |
0.0676971524 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 5 |
Hb_131646_010 |
0.0695963664 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_143395_010 |
0.0706022959 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 7 |
Hb_000256_190 |
0.0761700653 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 8 |
Hb_004631_180 |
0.0785094496 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 9 |
Hb_000576_020 |
0.0795461165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 10 |
Hb_004785_100 |
0.0797059148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007416_300 |
0.0798629554 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 12 |
Hb_002686_250 |
0.081525558 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_000928_220 |
0.0826715424 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 14 |
Hb_001077_030 |
0.082860194 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002311_070 |
0.0831605422 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 16 |
Hb_003029_030 |
0.0839778497 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006916_040 |
0.0860535502 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 18 |
Hb_144166_010 |
0.0866772016 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 19 |
Hb_000656_250 |
0.0869343314 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 20 |
Hb_000116_060 |
0.0871781966 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |