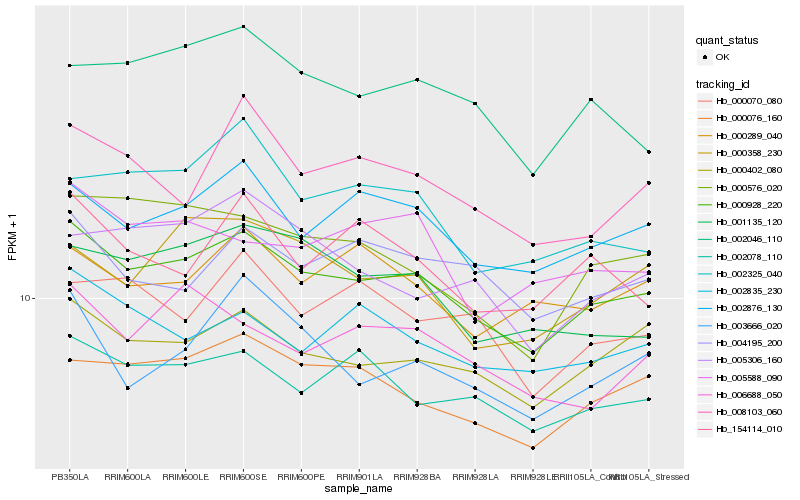
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_220 |
0.0 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 2 |
Hb_000402_080 |
0.0515910023 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 3 |
Hb_002876_130 |
0.05372777 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 4 |
Hb_000076_160 |
0.0552670058 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 5 |
Hb_002078_110 |
0.0594836871 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 6 |
Hb_002835_230 |
0.066996492 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002046_110 |
0.0671846272 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000576_020 |
0.0683204325 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 9 |
Hb_004195_200 |
0.0684182206 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 10 |
Hb_008103_060 |
0.068496023 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_000358_230 |
0.0694561968 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 12 |
Hb_001135_120 |
0.0709353845 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005588_090 |
0.0721209372 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 14 |
Hb_000289_040 |
0.0730000182 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 15 |
Hb_002325_040 |
0.0732777214 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006688_050 |
0.073462112 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003666_020 |
0.0738740381 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000070_080 |
0.0740879244 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005306_160 |
0.0741080995 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_154114_010 |
0.0741199884 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |