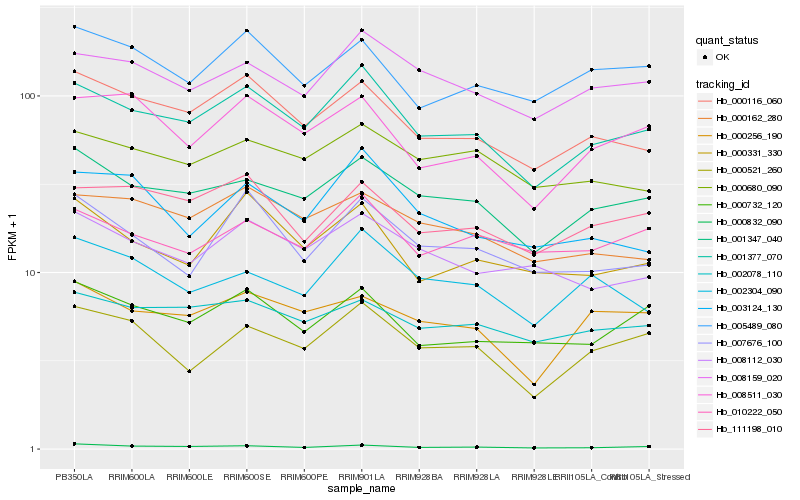
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001347_040 |
0.0644283491 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 3 |
Hb_000116_060 |
0.0673653013 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 4 |
Hb_008159_020 |
0.0752294929 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 5 |
Hb_111198_010 |
0.0836419153 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000732_120 |
0.0837747279 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008511_030 |
0.0840497137 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 8 |
Hb_000832_090 |
0.0846320261 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 9 |
Hb_000521_260 |
0.0847874091 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000162_280 |
0.0863587454 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 11 |
Hb_000331_330 |
0.087924018 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000680_090 |
0.0909134322 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 13 |
Hb_008112_030 |
0.0909455203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003124_130 |
0.0919110064 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002304_090 |
0.0920255104 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010222_050 |
0.0921406266 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000256_190 |
0.0921729943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 18 |
Hb_007676_100 |
0.0923343892 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 19 |
Hb_002078_110 |
0.0924743557 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 20 |
Hb_005489_080 |
0.0926477384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |