| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
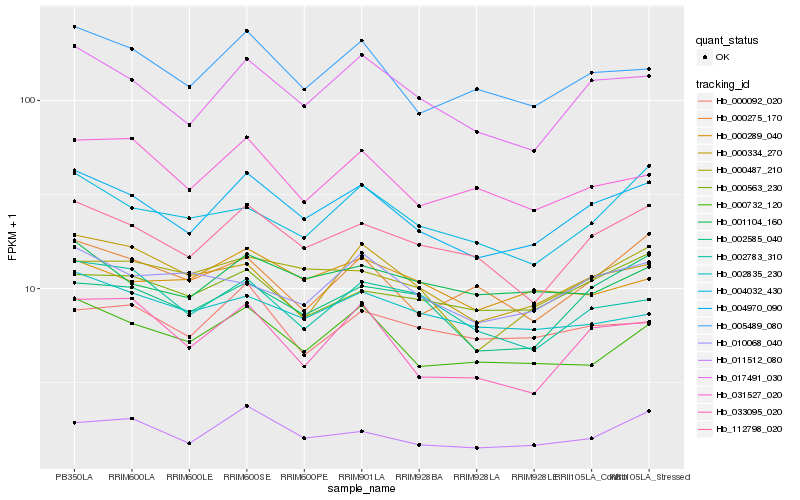
Hb_004970_090 |
0.0 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 2 |
Hb_017491_030 |
0.0514255285 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 3 |
Hb_112798_020 |
0.0620817198 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 4 |
Hb_005489_080 |
0.0697806084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000732_120 |
0.0779081335 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011512_080 |
0.0809295758 |
- |
- |
hypothetical protein JCGZ_06424 [Jatropha curcas] |
| 7 |
Hb_001104_160 |
0.083484497 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 8 |
Hb_002585_040 |
0.0858712125 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 9 |
Hb_000487_210 |
0.0870490802 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 10 |
Hb_031527_020 |
0.0873675951 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 11 |
Hb_000334_270 |
0.0877385186 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004032_430 |
0.0887645427 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 13 |
Hb_010068_040 |
0.0900326892 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 14 |
Hb_000563_230 |
0.0902790977 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 15 |
Hb_000275_170 |
0.0905802677 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 16 |
Hb_002783_310 |
0.0914393058 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 17 |
Hb_000289_040 |
0.0934332317 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 18 |
Hb_033095_020 |
0.0935417558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002835_230 |
0.0953801552 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000092_020 |
0.0959267207 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |