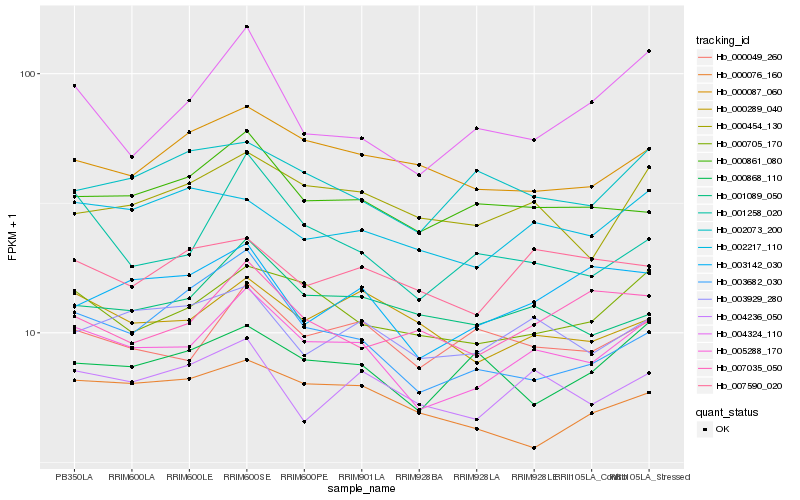
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 2 |
Hb_000705_170 |
0.0726735652 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 3 |
Hb_000049_260 |
0.080629313 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_004236_050 |
0.0844505924 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 5 |
Hb_000861_080 |
0.0850020614 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 6 |
Hb_003929_280 |
0.0850717084 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 7 |
Hb_000454_130 |
0.0862565755 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001089_050 |
0.0864878397 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_001258_020 |
0.0876058443 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 10 |
Hb_003142_030 |
0.088665066 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000289_040 |
0.0889045977 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 12 |
Hb_002217_110 |
0.0889466669 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 13 |
Hb_000087_060 |
0.0929370724 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_000868_110 |
0.0944994487 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 15 |
Hb_002073_200 |
0.0948486096 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 16 |
Hb_007590_020 |
0.0949580357 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 17 |
Hb_000076_160 |
0.0964188185 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 18 |
Hb_003682_030 |
0.0964647816 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 19 |
Hb_004324_110 |
0.0970159871 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 20 |
Hb_007035_050 |
0.0971848612 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |