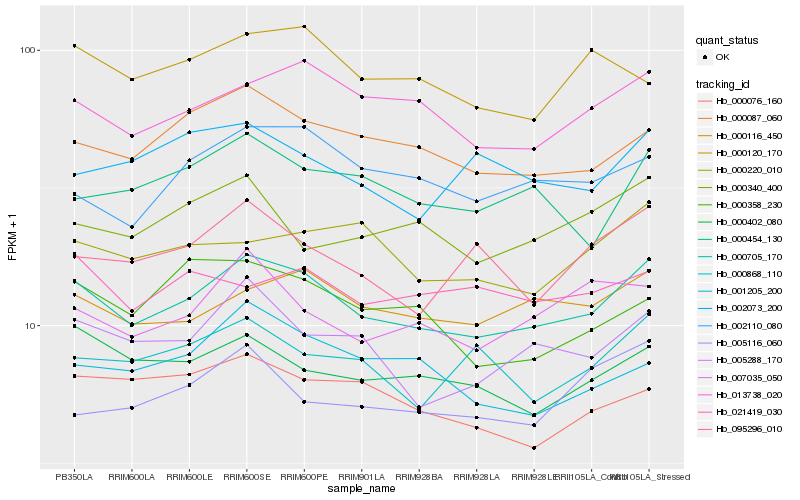
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 2 |
Hb_013738_020 |
0.0667102498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 3 |
Hb_000116_450 |
0.0677999323 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_000087_060 |
0.0692649448 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 5 |
Hb_005288_170 |
0.0726735652 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08610 [Populus euphratica] |
| 6 |
Hb_000402_080 |
0.0768813089 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 7 |
Hb_002110_080 |
0.0785632747 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 8 |
Hb_007035_050 |
0.0793432249 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 9 |
Hb_000220_010 |
0.0795626443 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 10 |
Hb_095296_010 |
0.0802589037 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 11 |
Hb_000454_130 |
0.0809019762 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000868_110 |
0.0814610503 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 13 |
Hb_000340_400 |
0.0824310074 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 14 |
Hb_021419_030 |
0.0858934873 |
- |
- |
hypothetical protein glysoja_023295 [Glycine soja] |
| 15 |
Hb_000076_160 |
0.0865942215 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 16 |
Hb_000120_170 |
0.0866091565 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 17 |
Hb_001205_200 |
0.086737147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002073_200 |
0.0874217799 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 19 |
Hb_000358_230 |
0.0878931441 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 20 |
Hb_005116_060 |
0.0883382761 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |