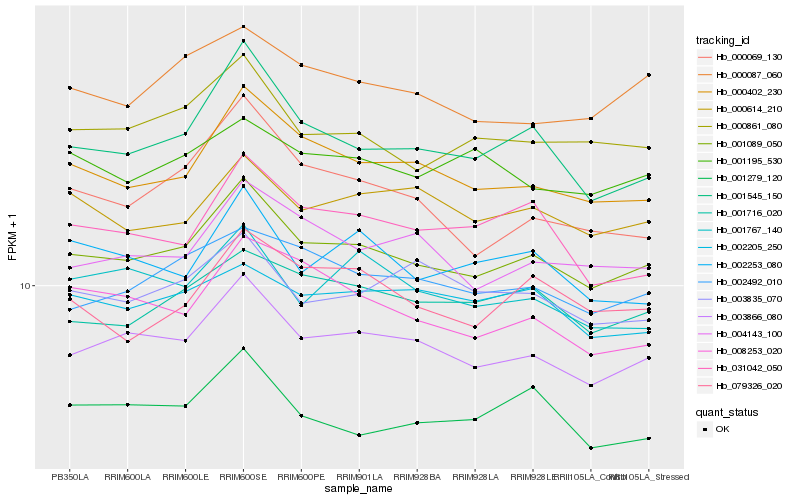
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_050 |
0.0 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_003866_080 |
0.0336894892 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 3 |
Hb_000402_230 |
0.0442840039 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000861_080 |
0.0490578084 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 5 |
Hb_001545_150 |
0.0506533758 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 6 |
Hb_004143_100 |
0.0547095922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001716_020 |
0.0557058086 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 8 |
Hb_002492_010 |
0.0564011205 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_000087_060 |
0.0572533148 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_000614_210 |
0.0596849956 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_079326_020 |
0.0616985351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_031042_050 |
0.0625731661 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 13 |
Hb_001195_530 |
0.0640673234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002253_080 |
0.0642102544 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 15 |
Hb_008253_020 |
0.0649245787 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 16 |
Hb_000069_130 |
0.065831039 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002205_250 |
0.0659377991 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001767_140 |
0.0660501987 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 19 |
Hb_001279_120 |
0.0660768255 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 20 |
Hb_003835_070 |
0.0662506882 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |