| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
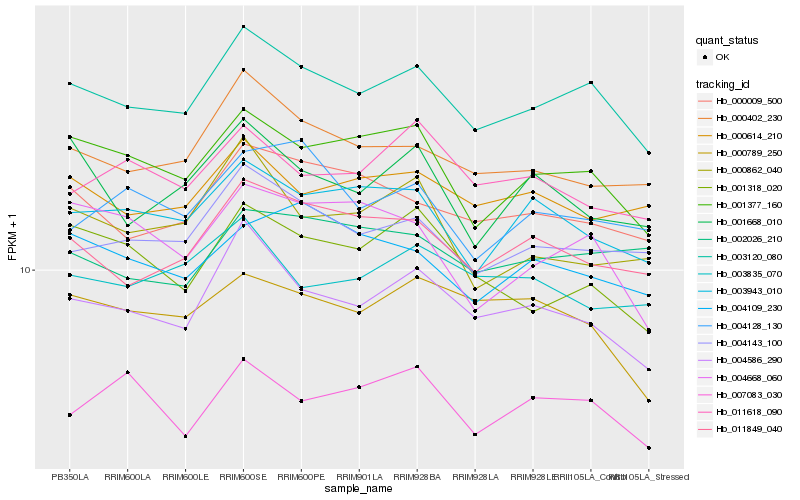
Hb_003120_080 |
0.0 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001377_160 |
0.0569427374 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 3 |
Hb_004586_290 |
0.0569668644 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 4 |
Hb_004143_100 |
0.0569763214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000402_230 |
0.0621614135 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_011849_040 |
0.0635545344 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000009_500 |
0.0655532145 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 8 |
Hb_003943_010 |
0.066844866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000614_210 |
0.0727378924 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002026_210 |
0.0768874875 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 11 |
Hb_004109_230 |
0.0783482814 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_001318_020 |
0.0796754934 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_000789_250 |
0.0804291472 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 14 |
Hb_004668_060 |
0.0807312491 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004128_130 |
0.0812377413 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001668_010 |
0.0814221208 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 17 |
Hb_000862_040 |
0.0818022616 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 18 |
Hb_007083_030 |
0.0822868475 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 19 |
Hb_011618_090 |
0.0823998334 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_003835_070 |
0.0824250331 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |