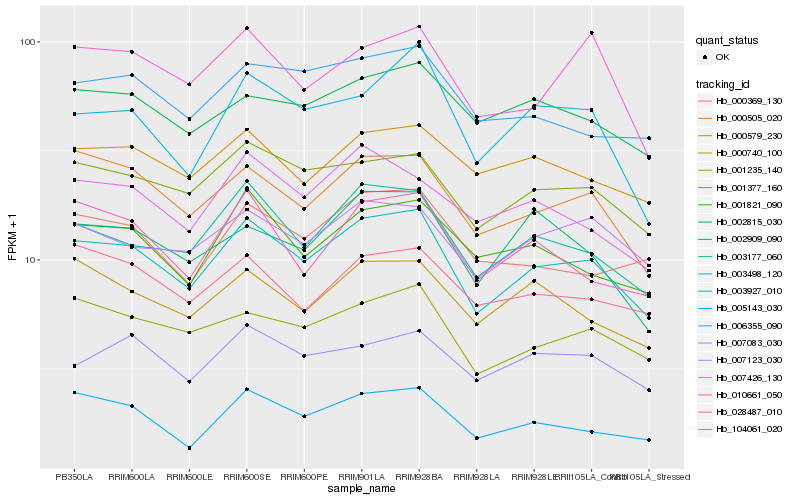
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003498_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 2 |
Hb_003177_060 |
0.0594675034 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001377_160 |
0.0649616192 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 4 |
Hb_000505_020 |
0.065025457 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 5 |
Hb_001235_140 |
0.0692019865 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 6 |
Hb_001821_090 |
0.0758627646 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 7 |
Hb_005143_030 |
0.0768908277 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 8 |
Hb_007123_030 |
0.07868632 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000740_100 |
0.0793161985 |
- |
- |
calpain, putative [Ricinus communis] |
| 10 |
Hb_007426_130 |
0.0805845469 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_028487_010 |
0.0807135046 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007083_030 |
0.0818718201 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 13 |
Hb_002815_030 |
0.0820507651 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 14 |
Hb_003927_010 |
0.0821418779 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_006355_090 |
0.082223338 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 16 |
Hb_000579_230 |
0.0827518196 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 17 |
Hb_010661_050 |
0.0848695087 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP]-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_104061_020 |
0.0849220971 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 19 |
Hb_000369_130 |
0.0850513519 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_002909_090 |
0.0851477207 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |