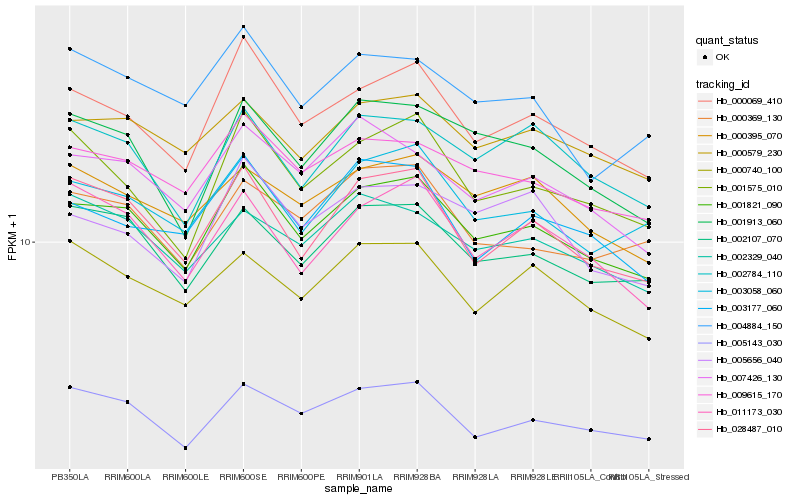
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001821_090 |
0.0 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 2 |
Hb_000069_410 |
0.0432589983 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 3 |
Hb_009615_170 |
0.05311677 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002107_070 |
0.0558954333 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 5 |
Hb_028487_010 |
0.0576164613 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003058_060 |
0.0587041601 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 7 |
Hb_000579_230 |
0.0620095356 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 8 |
Hb_007426_130 |
0.0644698801 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 9 |
Hb_001913_060 |
0.0656883328 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 10 |
Hb_001575_010 |
0.0676362599 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002784_110 |
0.0686838779 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 12 |
Hb_003177_060 |
0.0698168789 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000369_130 |
0.0699625648 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 14 |
Hb_000395_070 |
0.0724746578 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011173_030 |
0.0731102654 |
- |
- |
pumilio, putative [Ricinus communis] |
| 16 |
Hb_005143_030 |
0.0733845383 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 17 |
Hb_004884_150 |
0.0740379778 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 18 |
Hb_002329_040 |
0.0741224556 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 19 |
Hb_000740_100 |
0.0744362266 |
- |
- |
calpain, putative [Ricinus communis] |
| 20 |
Hb_005656_040 |
0.0751877091 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |