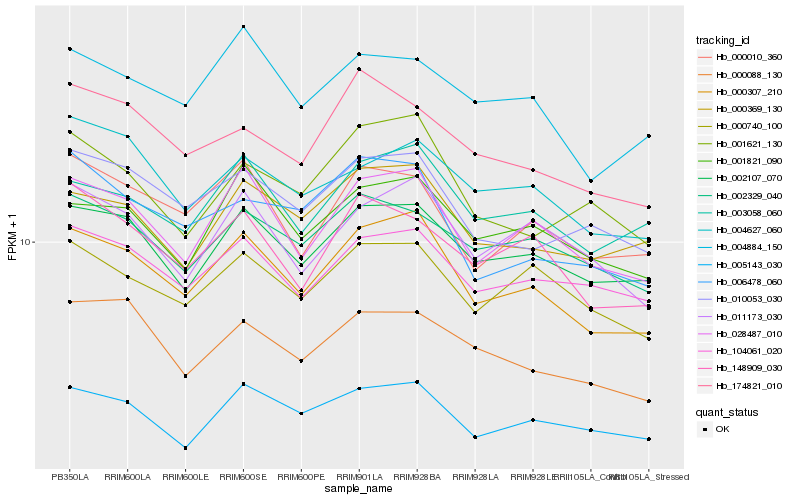
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_210 |
0.0 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_028487_010 |
0.0504435908 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002107_070 |
0.0671482956 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 4 |
Hb_003058_060 |
0.0737254455 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 5 |
Hb_011173_030 |
0.0742650134 |
- |
- |
pumilio, putative [Ricinus communis] |
| 6 |
Hb_010053_030 |
0.074775992 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 7 |
Hb_000740_100 |
0.0759650145 |
- |
- |
calpain, putative [Ricinus communis] |
| 8 |
Hb_000010_360 |
0.0761700282 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 9 |
Hb_104061_020 |
0.0777844259 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 10 |
Hb_001821_090 |
0.0779301974 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 11 |
Hb_000369_130 |
0.0785989035 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 12 |
Hb_005143_030 |
0.0787397066 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 13 |
Hb_148909_030 |
0.0818203683 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004884_150 |
0.0822490984 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 15 |
Hb_000088_130 |
0.0834588273 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 16 |
Hb_001621_130 |
0.083594363 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_006478_060 |
0.0845275618 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002329_040 |
0.0865035651 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 19 |
Hb_174821_010 |
0.0879798671 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 20 |
Hb_004627_060 |
0.0888782888 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |