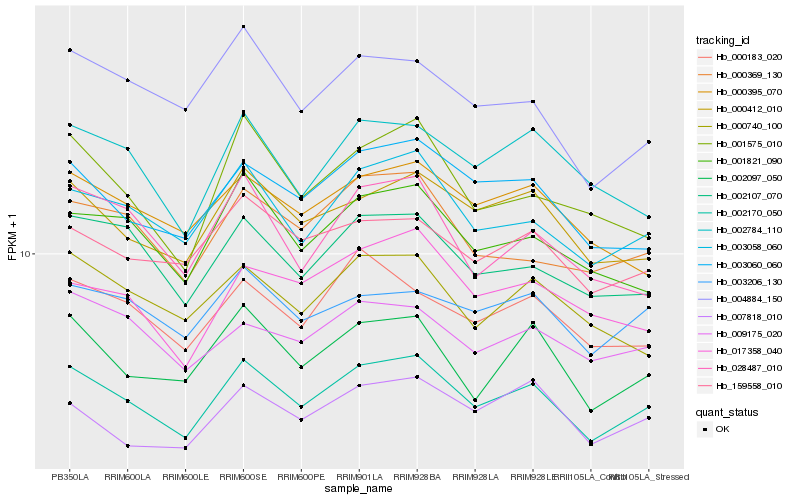
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002170_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |
| 2 |
Hb_003058_060 |
0.0648333962 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 3 |
Hb_000412_010 |
0.0687837923 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_002097_050 |
0.0720174954 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 5 |
Hb_003060_060 |
0.0729440539 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 6 |
Hb_002107_070 |
0.0773577107 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 7 |
Hb_001575_010 |
0.0793303448 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001821_090 |
0.0801413515 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 9 |
Hb_000369_130 |
0.0810489003 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_002784_110 |
0.0823885614 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 11 |
Hb_159558_010 |
0.0840862692 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 12 |
Hb_000183_020 |
0.0855225216 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007818_010 |
0.0860990453 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_003206_130 |
0.0861078466 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 15 |
Hb_004884_150 |
0.0866538006 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 16 |
Hb_028487_010 |
0.0866642834 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 17 |
Hb_017358_040 |
0.0884830958 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 18 |
Hb_000740_100 |
0.0886301918 |
- |
- |
calpain, putative [Ricinus communis] |
| 19 |
Hb_009175_020 |
0.0887084909 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000395_070 |
0.0900310185 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |