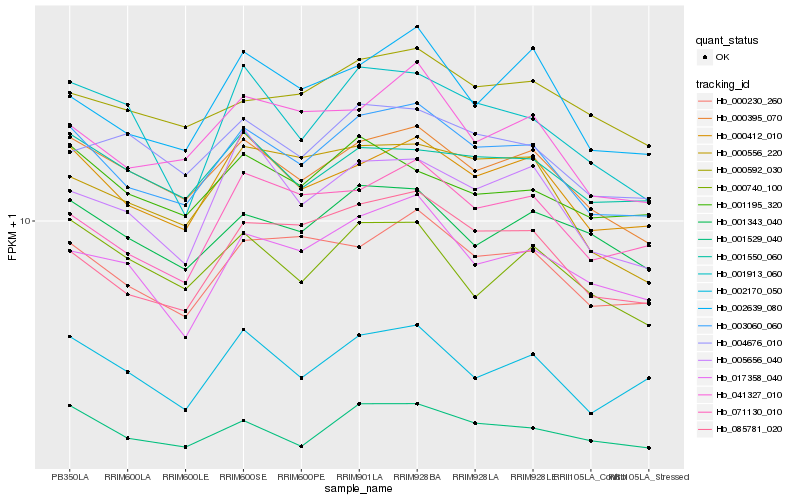
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003060_060 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 2 |
Hb_000412_010 |
0.0513439272 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000395_070 |
0.0522740693 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000230_260 |
0.0613966087 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 5 |
Hb_001529_040 |
0.0673181019 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 6 |
Hb_085781_020 |
0.0678609202 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_041327_010 |
0.0683026462 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 8 |
Hb_071130_010 |
0.0707684149 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_017358_040 |
0.0720362271 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 10 |
Hb_002170_050 |
0.0729440539 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |
| 11 |
Hb_001343_040 |
0.0735298643 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 12 |
Hb_002639_080 |
0.0742435746 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000556_220 |
0.0744879808 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 14 |
Hb_000592_030 |
0.0753502319 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 15 |
Hb_004676_010 |
0.0763555296 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_001550_060 |
0.0773234683 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000740_100 |
0.0801572173 |
- |
- |
calpain, putative [Ricinus communis] |
| 18 |
Hb_001195_320 |
0.0803946609 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 19 |
Hb_005656_040 |
0.0804863235 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_001913_060 |
0.0807016587 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |