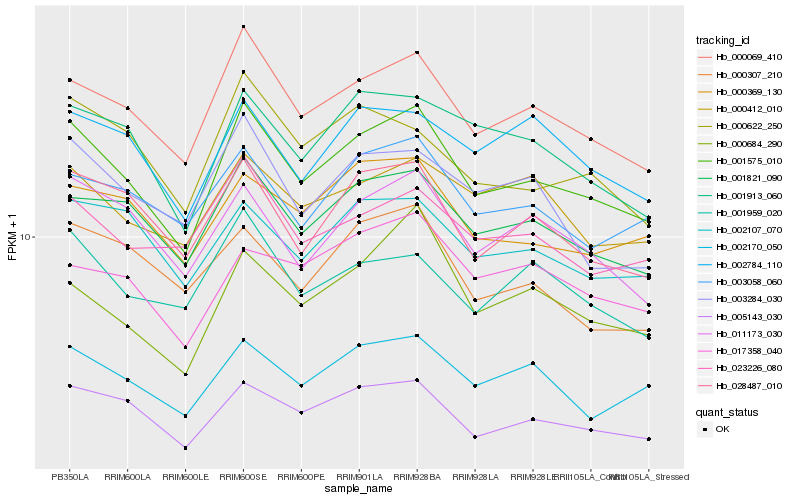
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001575_010 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000069_410 |
0.0544392732 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 3 |
Hb_005143_030 |
0.0631172821 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 4 |
Hb_001821_090 |
0.0676362599 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_028487_010 |
0.0759106111 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002170_050 |
0.0793303448 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |
| 7 |
Hb_011173_030 |
0.0807079367 |
- |
- |
pumilio, putative [Ricinus communis] |
| 8 |
Hb_001913_060 |
0.0825922105 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 9 |
Hb_000369_130 |
0.0845501441 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_002107_070 |
0.0852587155 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 11 |
Hb_003058_060 |
0.0859193676 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 12 |
Hb_023226_080 |
0.0889197379 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000412_010 |
0.0903450111 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001959_020 |
0.0907577684 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002784_110 |
0.0912860597 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 16 |
Hb_000622_250 |
0.0915202456 |
- |
- |
atpob1, putative [Ricinus communis] |
| 17 |
Hb_000684_290 |
0.0915463452 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 18 |
Hb_003284_030 |
0.0922792597 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_017358_040 |
0.0945766354 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 20 |
Hb_000307_210 |
0.0951050787 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |