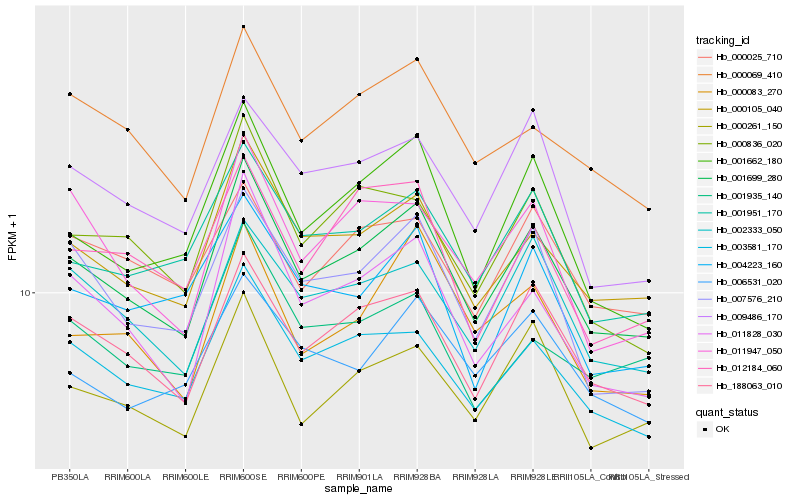
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001699_280 |
0.0 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 2 |
Hb_000105_040 |
0.0476890025 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 3 |
Hb_188063_010 |
0.0502744803 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 4 |
Hb_003581_170 |
0.0575966012 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000261_150 |
0.0618531515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001662_180 |
0.0634685104 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 7 |
Hb_001951_170 |
0.0652438394 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 8 |
Hb_002333_050 |
0.0656860876 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011828_030 |
0.0667412447 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 10 |
Hb_007576_210 |
0.0694005111 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 11 |
Hb_000069_410 |
0.0696035411 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 12 |
Hb_000836_020 |
0.0707006011 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 13 |
Hb_009486_170 |
0.0722832664 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 14 |
Hb_011947_050 |
0.0759300625 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 15 |
Hb_000083_270 |
0.0760522929 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000025_710 |
0.079029183 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 17 |
Hb_012184_060 |
0.0794564321 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006531_020 |
0.079520683 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 19 |
Hb_001935_140 |
0.0803567352 |
- |
- |
CCR4-NOT transcription complex subunit 1 [Gossypium arboreum] |
| 20 |
Hb_004223_160 |
0.0807072929 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |