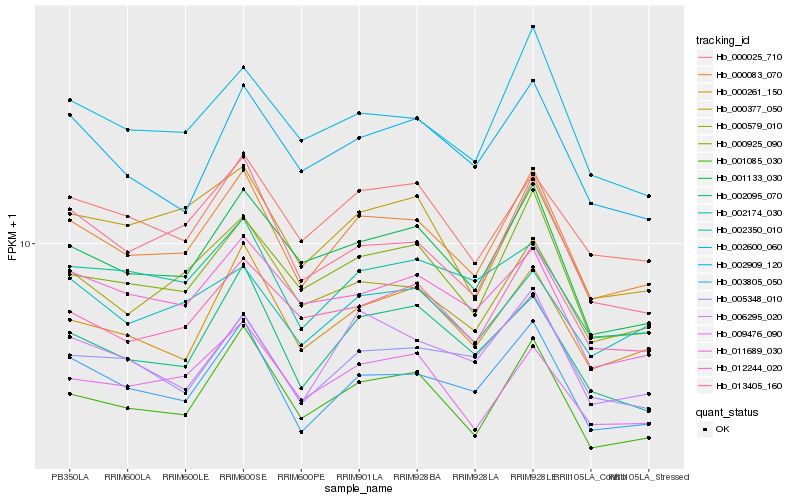
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_070 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001133_030 |
0.0606356602 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 3 |
Hb_000925_090 |
0.0608588003 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003805_050 |
0.0630323009 |
- |
- |
- |
| 5 |
Hb_000261_150 |
0.0644507258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000025_710 |
0.0659071379 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_012244_020 |
0.0679095286 |
- |
- |
calpain, putative [Ricinus communis] |
| 8 |
Hb_002600_060 |
0.0693664863 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 9 |
Hb_000579_010 |
0.0698192813 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 10 |
Hb_002909_120 |
0.0738140131 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 11 |
Hb_002350_010 |
0.0756197034 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 12 |
Hb_013405_160 |
0.0756628105 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000377_050 |
0.0764537884 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_011689_030 |
0.0766493767 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 15 |
Hb_005348_010 |
0.0768702688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009476_090 |
0.0775022662 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 17 |
Hb_006295_020 |
0.0777089289 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 18 |
Hb_002174_030 |
0.0777185974 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 19 |
Hb_002095_070 |
0.0791822598 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 20 |
Hb_001085_030 |
0.0792058756 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |