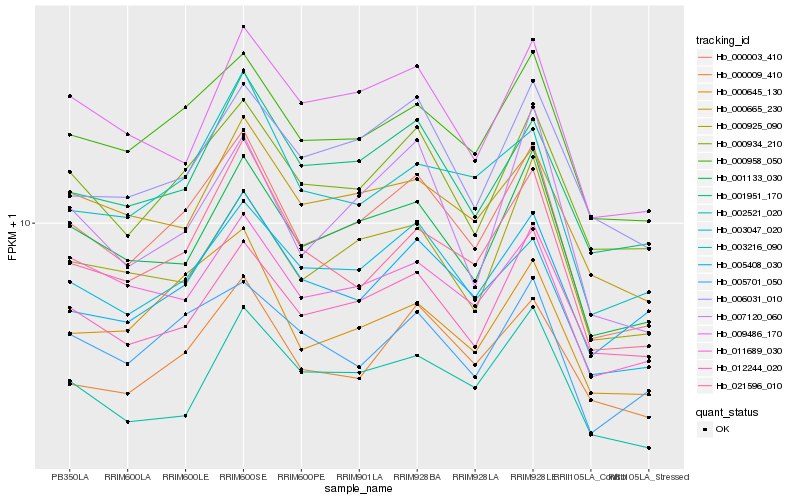
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_410 |
0.0 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 2 |
Hb_000958_050 |
0.0557593507 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 3 |
Hb_001133_030 |
0.0601394183 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 4 |
Hb_000934_210 |
0.0633501927 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 5 |
Hb_003216_090 |
0.0647838944 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 6 |
Hb_021596_010 |
0.0676377327 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 7 |
Hb_000009_410 |
0.0724476984 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 8 |
Hb_000645_130 |
0.072989126 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 9 |
Hb_003047_020 |
0.0732148702 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001951_170 |
0.0735582889 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_005701_050 |
0.0750109707 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 12 |
Hb_011689_030 |
0.0764939517 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 13 |
Hb_000665_230 |
0.0780306538 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 14 |
Hb_009486_170 |
0.0802564171 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 15 |
Hb_000925_090 |
0.0806936098 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005408_030 |
0.0808378292 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 17 |
Hb_002521_020 |
0.0816094795 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_007120_060 |
0.0820307779 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 19 |
Hb_006031_010 |
0.0827696226 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 20 |
Hb_012244_020 |
0.0828959546 |
- |
- |
calpain, putative [Ricinus communis] |