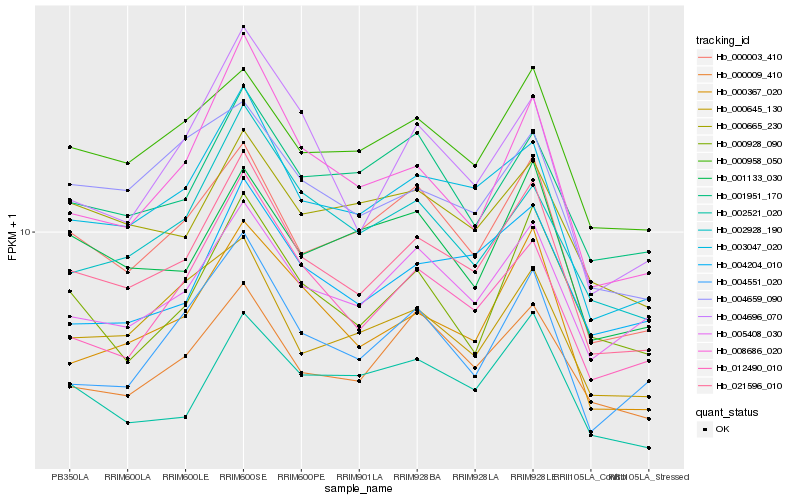
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021596_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 2 |
Hb_003047_020 |
0.0510105041 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000367_020 |
0.0655038456 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 4 |
Hb_000003_410 |
0.0676377327 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 5 |
Hb_000958_050 |
0.0743353575 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 6 |
Hb_005408_030 |
0.0758866754 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 7 |
Hb_008686_020 |
0.0774489729 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012490_010 |
0.0779102377 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000009_410 |
0.0785014391 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 10 |
Hb_004551_020 |
0.0812402241 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 11 |
Hb_002521_020 |
0.0830766656 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_004204_010 |
0.0834829289 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 13 |
Hb_000928_090 |
0.0848318228 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 14 |
Hb_000645_130 |
0.085249767 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_000665_230 |
0.0861249671 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 16 |
Hb_002928_190 |
0.0862983393 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 17 |
Hb_004696_070 |
0.087087151 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 18 |
Hb_001951_170 |
0.0871613645 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_004659_090 |
0.0875107013 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 20 |
Hb_001133_030 |
0.087515808 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |