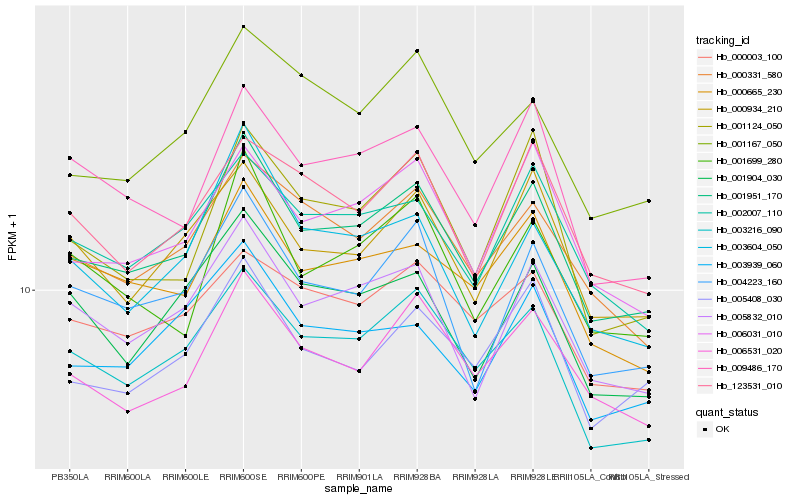
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 2 |
Hb_003216_090 |
0.051403809 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005832_010 |
0.0545175932 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006031_010 |
0.0563394493 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 5 |
Hb_006531_020 |
0.0568318404 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 6 |
Hb_002007_110 |
0.0595736604 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000331_580 |
0.0595779754 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 8 |
Hb_004223_160 |
0.0601392183 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005408_030 |
0.0610621851 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 10 |
Hb_000665_230 |
0.0620015735 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 11 |
Hb_000934_210 |
0.0620135896 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 12 |
Hb_009486_170 |
0.0629992121 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 13 |
Hb_001699_280 |
0.0652438394 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 14 |
Hb_123531_010 |
0.0664266402 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 15 |
Hb_003939_060 |
0.066859343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001167_050 |
0.0676198759 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 17 |
Hb_001904_030 |
0.0676771858 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 18 |
Hb_003604_050 |
0.0678003566 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001124_050 |
0.0680877055 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 20 |
Hb_000003_100 |
0.0689678069 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |