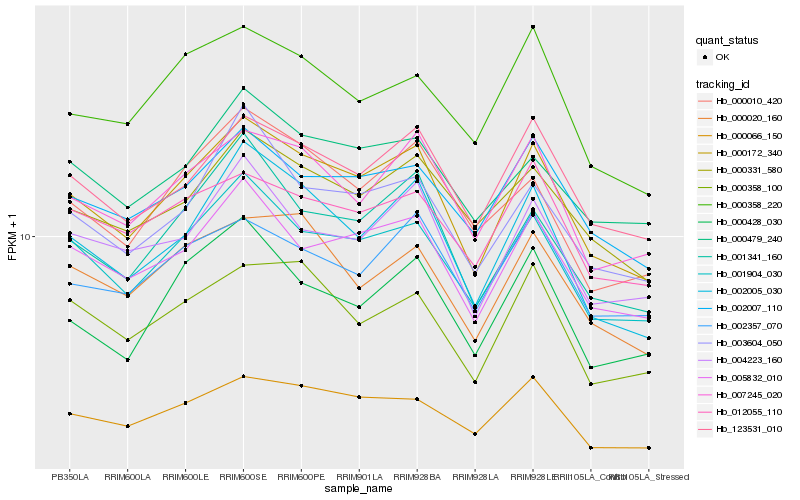
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 2 |
Hb_001904_030 |
0.0486851956 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 3 |
Hb_005832_010 |
0.0565852473 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002005_030 |
0.0578141849 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 5 |
Hb_123531_010 |
0.0595116897 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 6 |
Hb_007245_020 |
0.0628083587 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000066_150 |
0.064982908 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 8 |
Hb_001341_160 |
0.0666396199 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002007_110 |
0.066943092 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004223_160 |
0.0691669812 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000331_580 |
0.0702075696 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_000020_160 |
0.0723064902 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 13 |
Hb_000479_240 |
0.0736706331 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003604_050 |
0.0738688938 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000428_030 |
0.0741383793 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 16 |
Hb_000358_100 |
0.0741421127 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 17 |
Hb_000010_420 |
0.0757375675 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 18 |
Hb_000358_220 |
0.0766709209 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 19 |
Hb_012055_110 |
0.0772383251 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 20 |
Hb_002357_070 |
0.0777420692 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |