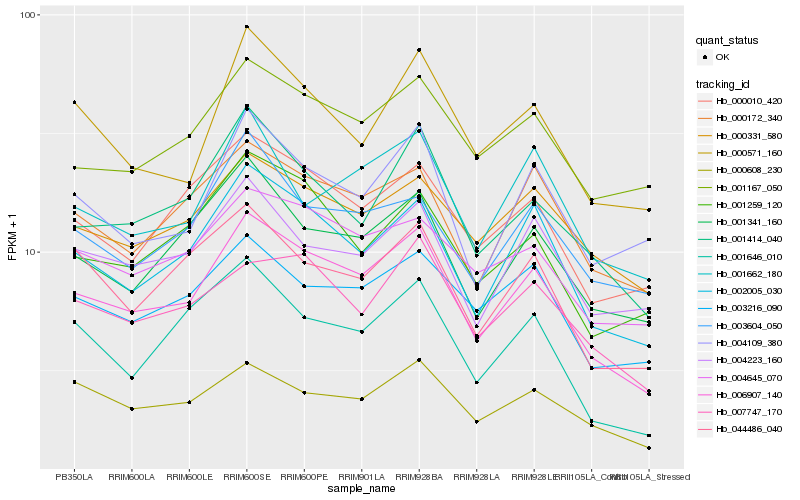
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006907_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002005_030 |
0.0603576386 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 3 |
Hb_001414_040 |
0.0787369559 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001341_160 |
0.0803934318 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000172_340 |
0.0808872958 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 6 |
Hb_000010_420 |
0.0829154451 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 7 |
Hb_004645_070 |
0.0848824461 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 8 |
Hb_007747_170 |
0.0862934428 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 9 |
Hb_000331_580 |
0.0876007384 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 10 |
Hb_001259_120 |
0.0877466322 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 11 |
Hb_001167_050 |
0.0879838808 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 12 |
Hb_003216_090 |
0.0888037476 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001646_010 |
0.0890655842 |
- |
- |
hypothetical protein JCGZ_16333 [Jatropha curcas] |
| 14 |
Hb_004223_160 |
0.0893624908 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_044486_040 |
0.089402041 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 16 |
Hb_000571_160 |
0.0910059284 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 17 |
Hb_003604_050 |
0.0918251468 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004109_380 |
0.0918643133 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 19 |
Hb_001662_180 |
0.0931600532 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 20 |
Hb_000608_230 |
0.0937246889 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |