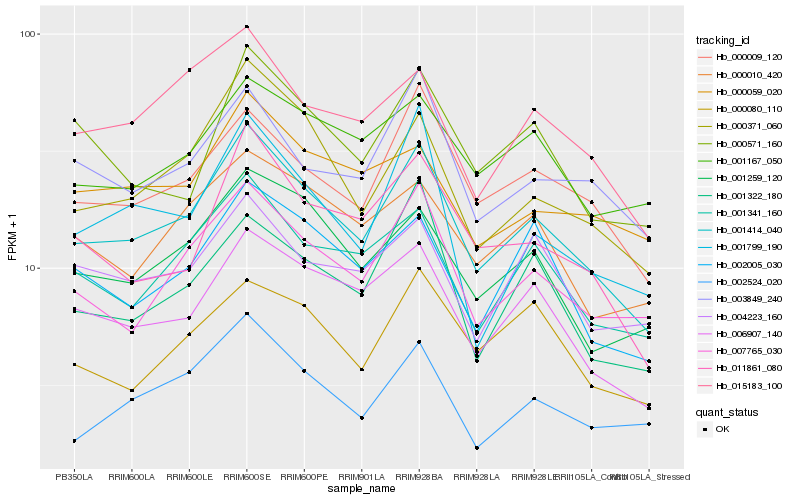
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001414_040 |
0.0 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_006907_140 |
0.0787369559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000371_060 |
0.08124374 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 4 |
Hb_007765_030 |
0.0814538002 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 5 |
Hb_000009_120 |
0.0817664262 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 6 |
Hb_001341_160 |
0.0836082891 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001259_120 |
0.089368909 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 8 |
Hb_011861_080 |
0.0943401221 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 9 |
Hb_015183_100 |
0.0950461532 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 10 |
Hb_002005_030 |
0.0972517465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_003849_240 |
0.0977433973 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000010_420 |
0.0993125161 |
- |
- |
Callose synthase 10 [Morus notabilis] |
| 13 |
Hb_002524_020 |
0.1001000002 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 14 |
Hb_001167_050 |
0.1023040448 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 15 |
Hb_001322_180 |
0.1025063078 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 16 |
Hb_000571_160 |
0.1036100661 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 17 |
Hb_000080_110 |
0.1052884707 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004223_160 |
0.1064817663 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001799_190 |
0.107619504 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 20 |
Hb_000059_020 |
0.1078337525 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |