| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
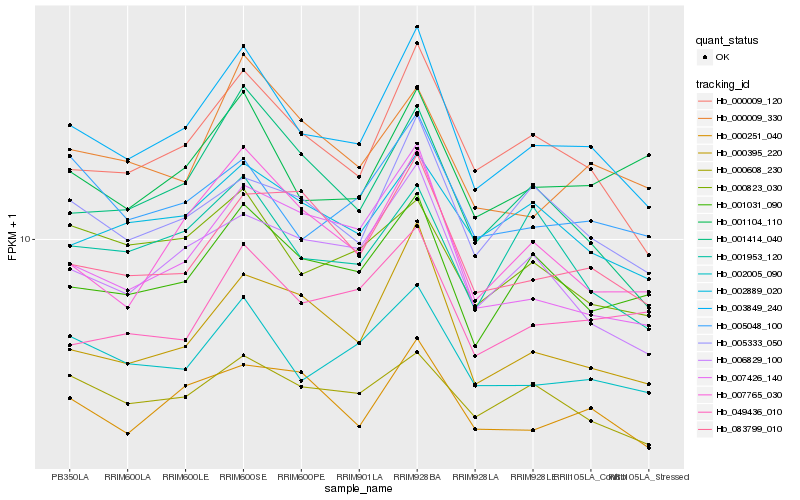
Hb_003849_240 |
0.0 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000009_120 |
0.0687556165 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 3 |
Hb_002005_090 |
0.0823304809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001031_090 |
0.0917938261 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 5 |
Hb_007765_030 |
0.0925855815 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 6 |
Hb_000395_220 |
0.097444813 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_005333_050 |
0.0974560319 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006829_100 |
0.0974847672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001414_040 |
0.0977433973 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007426_140 |
0.1056829501 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_002889_020 |
0.1057812697 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 12 |
Hb_000608_230 |
0.1065635963 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 13 |
Hb_005048_100 |
0.106751753 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 14 |
Hb_000251_040 |
0.1067577116 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 15 |
Hb_083799_010 |
0.1076190701 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_001104_110 |
0.1076478329 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Populus euphratica] |
| 17 |
Hb_000823_030 |
0.1093878009 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 18 |
Hb_001953_120 |
0.1096807781 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000009_330 |
0.1110385307 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_049436_010 |
0.1128378262 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |