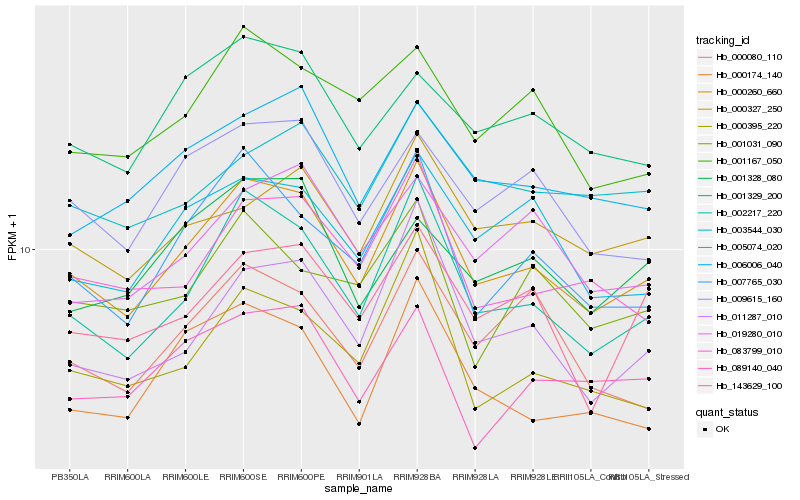
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_660 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002217_220 |
0.0569357202 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 3 |
Hb_007765_030 |
0.0804209503 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 4 |
Hb_083799_010 |
0.087006569 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_009615_160 |
0.0888254197 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 6 |
Hb_005074_020 |
0.0905372623 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006006_040 |
0.0928251755 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 8 |
Hb_011287_010 |
0.0933654123 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000327_250 |
0.0949154833 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 10 |
Hb_143629_100 |
0.0949507353 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 11 |
Hb_001167_050 |
0.0949636288 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 12 |
Hb_001329_200 |
0.0968013695 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 13 |
Hb_019280_010 |
0.0989947191 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000080_110 |
0.1022215037 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003544_030 |
0.103562884 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 16 |
Hb_000395_220 |
0.1039662034 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_089140_040 |
0.1042191326 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 18 |
Hb_000174_140 |
0.1044309868 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 19 |
Hb_001031_090 |
0.1049243732 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 20 |
Hb_001328_080 |
0.1054322241 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |