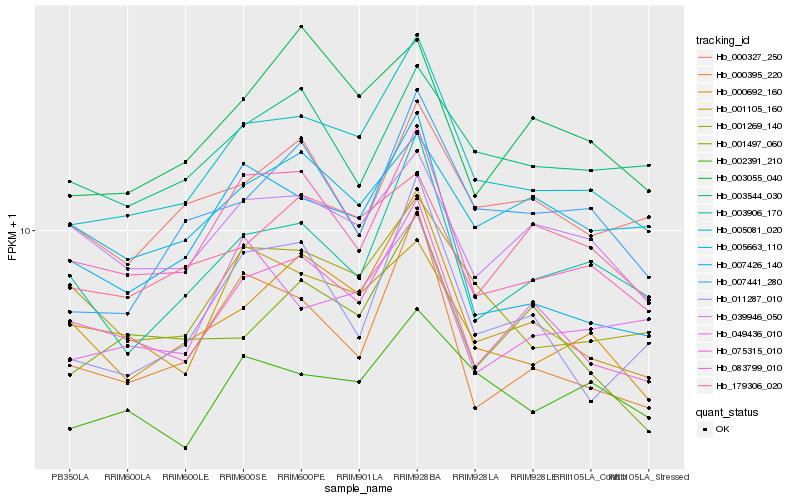
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005081_020 |
0.0 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 2 |
Hb_083799_010 |
0.0818550334 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000692_160 |
0.0832743128 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 4 |
Hb_075315_010 |
0.0859163291 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 5 |
Hb_002391_210 |
0.0903696415 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 6 |
Hb_001269_140 |
0.0904616848 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 7 |
Hb_003906_170 |
0.0922163215 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_007441_280 |
0.0988042088 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 9 |
Hb_001105_160 |
0.1035369589 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 10 |
Hb_039946_050 |
0.1047136047 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000395_220 |
0.1053265811 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_007426_140 |
0.1062972511 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 13 |
Hb_005663_110 |
0.1069941073 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003544_030 |
0.1071135254 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 15 |
Hb_000327_250 |
0.1071276503 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 16 |
Hb_049436_010 |
0.1076568014 |
- |
- |
PREDICTED: UPF0202 protein At1g10490-like [Glycine max] |
| 17 |
Hb_001497_060 |
0.1087642426 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_179306_020 |
0.1090747226 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003055_040 |
0.1094736289 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_011287_010 |
0.1098960709 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |