| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
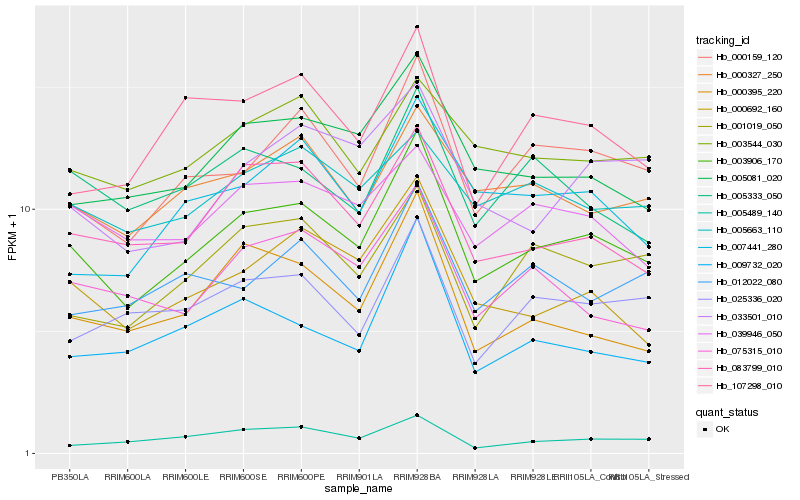
Hb_003906_170 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000159_120 |
0.0823793176 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 3 |
Hb_000692_160 |
0.0921561765 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 4 |
Hb_005081_020 |
0.0922163215 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 5 |
Hb_083799_010 |
0.0946481254 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000327_250 |
0.10219388 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 7 |
Hb_075315_010 |
0.1036687589 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 8 |
Hb_000395_220 |
0.1038516922 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_005333_050 |
0.1053599206 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001019_050 |
0.1066346063 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 11 |
Hb_009732_020 |
0.1075293938 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 12 |
Hb_107298_010 |
0.1076620622 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 13 |
Hb_039946_050 |
0.1078532442 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_025336_020 |
0.1089280231 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 15 |
Hb_012022_080 |
0.11053891 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_003544_030 |
0.1119210041 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 17 |
Hb_007441_280 |
0.1125347748 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 18 |
Hb_033501_010 |
0.1135392631 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 19 |
Hb_005489_140 |
0.1145801114 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 20 |
Hb_005663_110 |
0.1154463901 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |