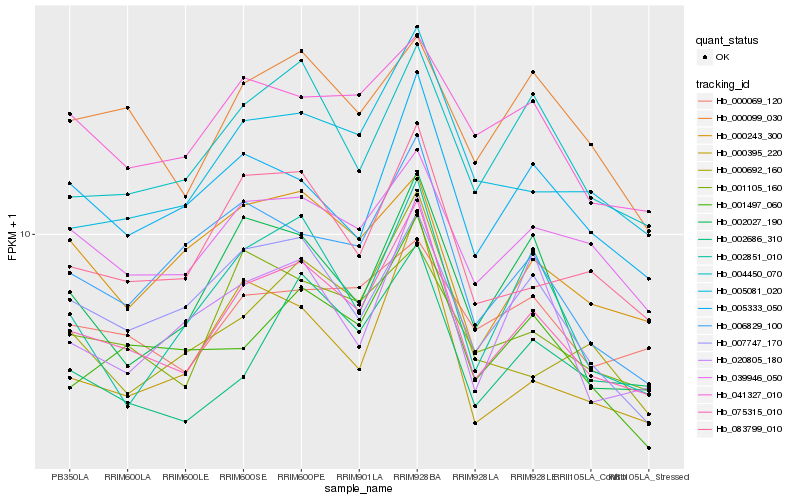
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075315_010 |
0.0 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 2 |
Hb_083799_010 |
0.0774919105 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_005081_020 |
0.0859163291 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 4 |
Hb_039946_050 |
0.0860585706 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_004450_070 |
0.0864062406 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 6 |
Hb_007747_170 |
0.0882397307 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 7 |
Hb_001105_160 |
0.0886019861 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 8 |
Hb_001497_060 |
0.0916118834 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 9 |
Hb_006829_100 |
0.0927669671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002686_310 |
0.0938652529 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 11 |
Hb_000395_220 |
0.0942206587 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_002027_190 |
0.0942390427 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 13 |
Hb_005333_050 |
0.0947843474 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_041327_010 |
0.0966498981 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 15 |
Hb_000099_030 |
0.0970067357 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_002851_010 |
0.0972507602 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_000243_300 |
0.0999066279 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000692_160 |
0.1001469068 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_020805_180 |
0.100361674 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 20 |
Hb_000069_120 |
0.100402829 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |