| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
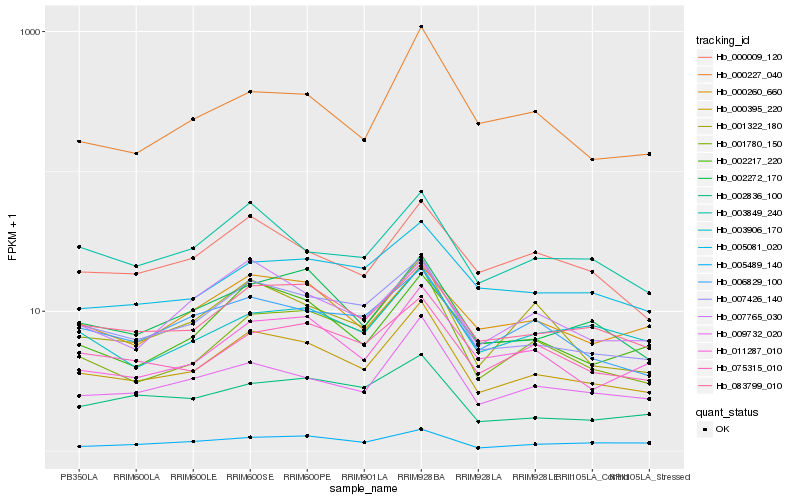
Hb_000395_220 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 2 |
Hb_007426_140 |
0.0648711933 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_083799_010 |
0.0729443539 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_009732_020 |
0.0830918341 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 5 |
Hb_001780_150 |
0.0885657633 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 6 |
Hb_002272_170 |
0.0915479545 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 7 |
Hb_011287_010 |
0.0918729247 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_002217_220 |
0.0922202974 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 9 |
Hb_075315_010 |
0.0942206587 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 10 |
Hb_001322_180 |
0.0951310869 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 11 |
Hb_006829_100 |
0.0961099312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003849_240 |
0.097444813 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000009_120 |
0.1011471561 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 14 |
Hb_007765_030 |
0.1027579341 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 15 |
Hb_003906_170 |
0.1038516922 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000260_660 |
0.1039662034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005081_020 |
0.1053265811 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 18 |
Hb_000227_040 |
0.1084185841 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 19 |
Hb_002836_100 |
0.1090327728 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005489_140 |
0.111471867 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |