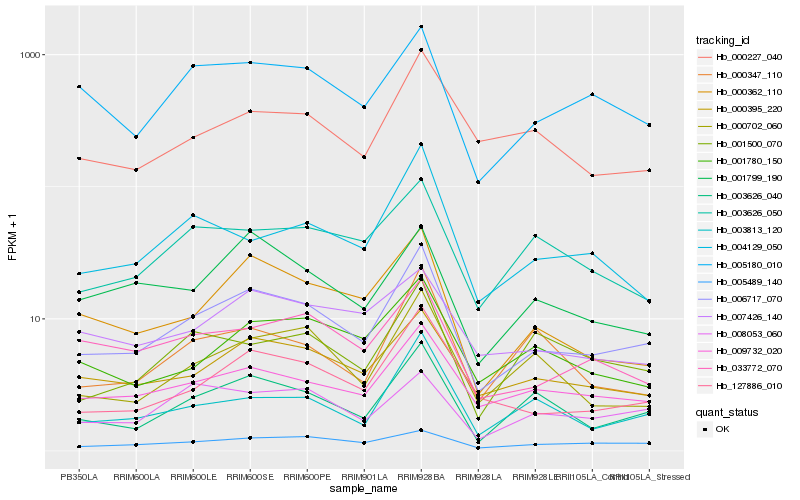
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006717_070 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 2 |
Hb_127886_010 |
0.1076630309 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_009732_020 |
0.1108213664 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 4 |
Hb_003626_040 |
0.112040133 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000395_220 |
0.1137479354 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_000362_110 |
0.1234403828 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 7 |
Hb_003813_120 |
0.1342972771 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_005180_010 |
0.1352014201 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 9 |
Hb_000702_060 |
0.1367850661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_033772_070 |
0.1375789699 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 11 |
Hb_000227_040 |
0.1376999438 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 12 |
Hb_004129_050 |
0.1393854619 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 13 |
Hb_001780_150 |
0.1398156828 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 14 |
Hb_005489_140 |
0.141970996 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 15 |
Hb_003626_050 |
0.1452562329 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 16 |
Hb_001799_190 |
0.1458462035 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 17 |
Hb_007426_140 |
0.1478893298 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 18 |
Hb_000347_110 |
0.1485649058 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 19 |
Hb_001500_070 |
0.1517546211 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008053_060 |
0.1524977425 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |