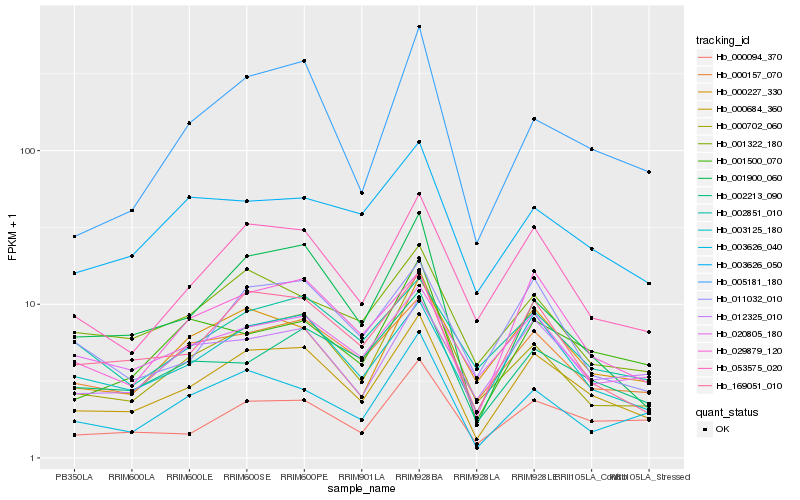
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_360 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 2 |
Hb_003125_180 |
0.0919039332 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 3 |
Hb_053575_020 |
0.0945061633 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 4 |
Hb_169051_010 |
0.097931557 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 5 |
Hb_000157_070 |
0.1093125939 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 6 |
Hb_000702_060 |
0.1117847825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_029879_120 |
0.1148376307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000094_370 |
0.1159332729 |
- |
- |
- |
| 9 |
Hb_000227_330 |
0.116863235 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 10 |
Hb_012325_010 |
0.1194353142 |
- |
- |
hypothetical protein RCOM_0068670 [Ricinus communis] |
| 11 |
Hb_002213_090 |
0.1206628765 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 12 |
Hb_011032_010 |
0.12267574 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 13 |
Hb_003626_040 |
0.1249845257 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001322_180 |
0.1267868984 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 15 |
Hb_001500_070 |
0.1304575574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005181_180 |
0.1305532617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001900_060 |
0.1307557254 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 18 |
Hb_002851_010 |
0.1336570652 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_003626_050 |
0.1339093553 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 20 |
Hb_020805_180 |
0.1343970344 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |