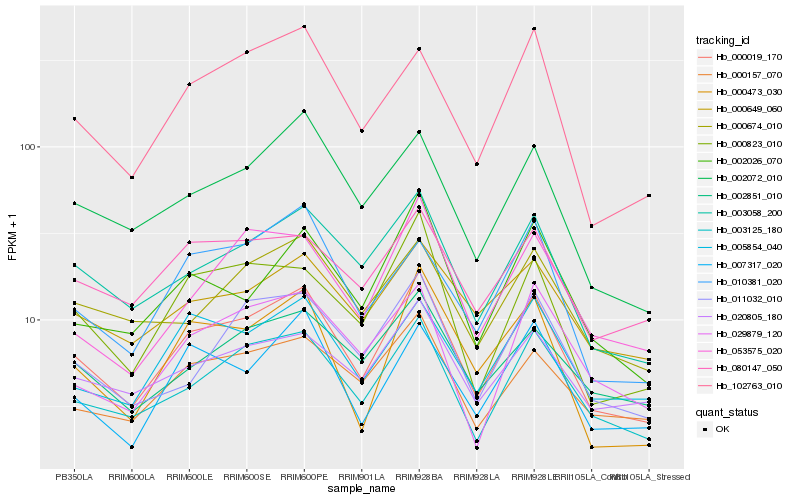
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_170 |
0.0 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 2 |
Hb_003125_180 |
0.077885294 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 3 |
Hb_003058_200 |
0.0804900678 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 4 |
Hb_002072_010 |
0.0970706135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_102763_010 |
0.0983511922 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 6 |
Hb_000823_010 |
0.1006829163 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000157_070 |
0.102366028 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 8 |
Hb_005854_040 |
0.1033234368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007317_020 |
0.104312641 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000473_030 |
0.1061892857 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 11 |
Hb_002851_010 |
0.1064406806 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 12 |
Hb_011032_010 |
0.1066094888 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 13 |
Hb_020805_180 |
0.1100186337 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 14 |
Hb_002026_070 |
0.1108829668 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 15 |
Hb_010381_020 |
0.1110989519 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 16 |
Hb_053575_020 |
0.1116371569 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 17 |
Hb_080147_050 |
0.1116870968 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 18 |
Hb_000674_010 |
0.1123126713 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000649_060 |
0.1127827706 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 20 |
Hb_029879_120 |
0.1142234205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |