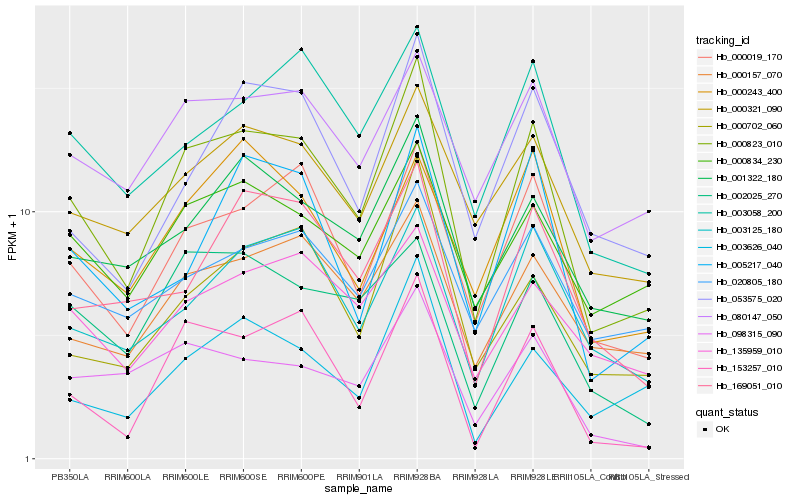
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000823_010 |
0.0 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 2 |
Hb_000019_170 |
0.1006829163 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 3 |
Hb_153257_010 |
0.1215989038 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 4 |
Hb_000157_070 |
0.1217202699 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 5 |
Hb_001322_180 |
0.1233087637 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 6 |
Hb_003058_200 |
0.1265113735 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 7 |
Hb_000834_230 |
0.1282096849 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 8 |
Hb_003125_180 |
0.1287760768 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 9 |
Hb_080147_050 |
0.1293329554 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 10 |
Hb_000321_090 |
0.1294614187 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 11 |
Hb_000702_060 |
0.13209879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_135959_010 |
0.1333125323 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 13 |
Hb_098315_090 |
0.1336586079 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 14 |
Hb_053575_020 |
0.1339450336 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 15 |
Hb_020805_180 |
0.1354440228 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 16 |
Hb_169051_010 |
0.1354452414 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 17 |
Hb_003626_040 |
0.1355436536 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002025_270 |
0.1371900939 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005217_040 |
0.1402417737 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000243_400 |
0.1408336604 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |