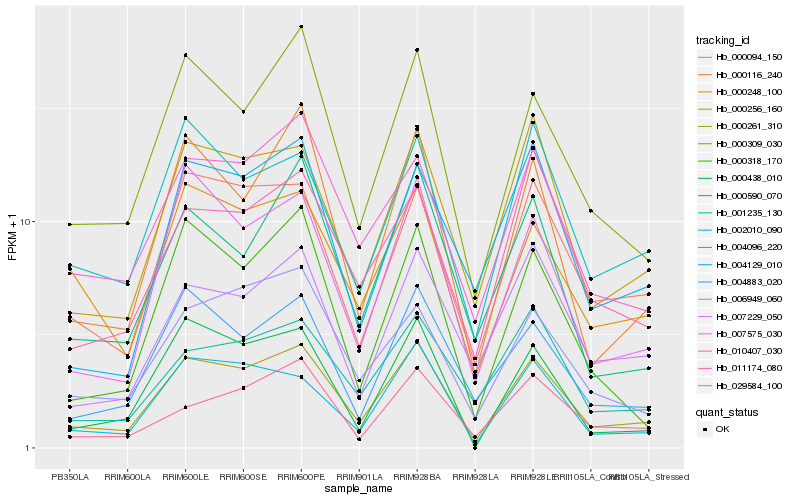
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_310 |
0.0 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 2 |
Hb_000256_160 |
0.0935358585 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 3 |
Hb_000309_030 |
0.0948939203 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000094_150 |
0.097339856 |
- |
- |
- |
| 5 |
Hb_000116_240 |
0.098830172 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 6 |
Hb_007575_030 |
0.1001376733 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 7 |
Hb_004096_220 |
0.1038342138 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010407_030 |
0.1119058474 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 9 |
Hb_004883_020 |
0.1144343643 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 10 |
Hb_002010_090 |
0.115031578 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_006949_060 |
0.1163142926 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 12 |
Hb_000248_100 |
0.1183039049 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 13 |
Hb_000438_010 |
0.1185888204 |
- |
- |
- |
| 14 |
Hb_000590_070 |
0.1202452221 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 15 |
Hb_011174_080 |
0.1211802075 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 16 |
Hb_000318_170 |
0.1211815787 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 17 |
Hb_007229_050 |
0.1242883215 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001235_130 |
0.1245587072 |
- |
- |
- |
| 19 |
Hb_004129_010 |
0.1247297064 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_029584_100 |
0.1275967337 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |