| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
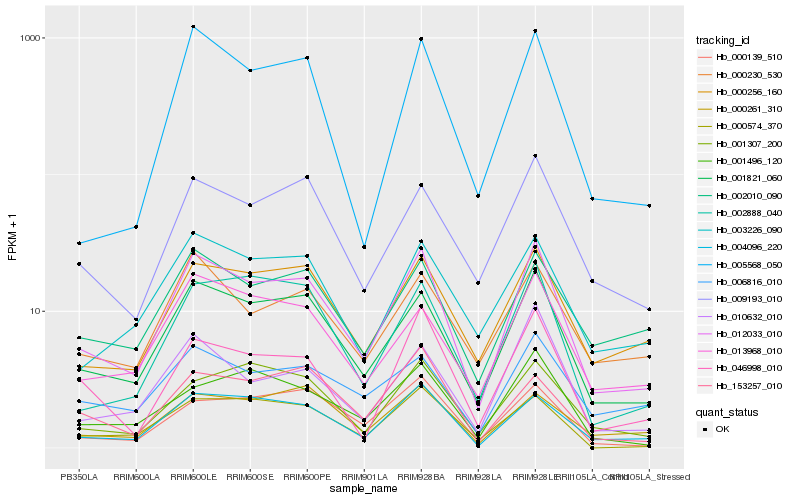
Hb_012033_010 |
0.0 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 2 |
Hb_001821_060 |
0.0726133146 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 3 |
Hb_004096_220 |
0.0992473823 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000256_160 |
0.1213100746 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 5 |
Hb_009193_010 |
0.1234442098 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 6 |
Hb_013968_010 |
0.129014521 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 7 |
Hb_002888_040 |
0.1311822251 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 8 |
Hb_005568_050 |
0.1334006592 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 9 |
Hb_000139_510 |
0.1353013677 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002010_090 |
0.1356741855 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001307_200 |
0.1357308264 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 12 |
Hb_003226_090 |
0.136012635 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 13 |
Hb_001496_120 |
0.1360904669 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000261_310 |
0.1365325435 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 15 |
Hb_010632_010 |
0.1372063576 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000574_370 |
0.1379037388 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 17 |
Hb_006816_010 |
0.1403993023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_153257_010 |
0.1405099462 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 19 |
Hb_046998_010 |
0.1414699297 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_000230_530 |
0.1421377367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |