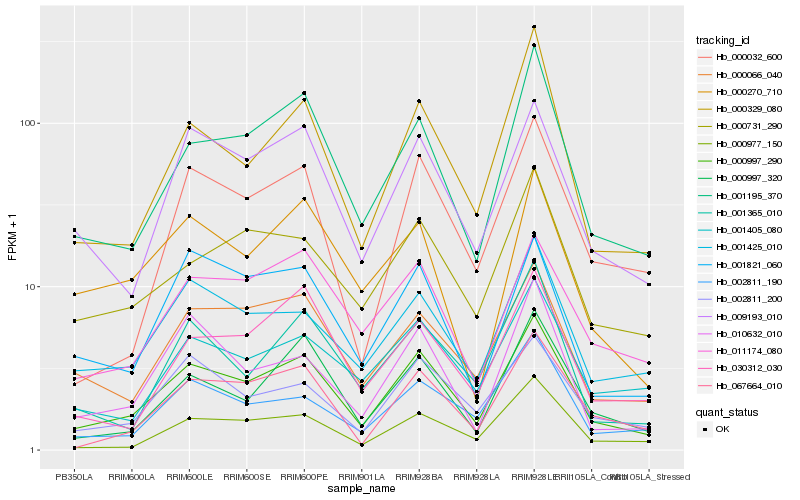
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_290 |
0.0 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 2 |
Hb_001195_370 |
0.0994463557 |
- |
- |
- |
| 3 |
Hb_009193_010 |
0.1083865168 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 4 |
Hb_000329_080 |
0.1113000045 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000066_040 |
0.115119496 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 6 |
Hb_001425_010 |
0.1151864985 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_011174_080 |
0.1197921098 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 8 |
Hb_002811_200 |
0.1198597566 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 9 |
Hb_000997_320 |
0.1207513415 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 10 |
Hb_067664_010 |
0.1217502249 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_000977_150 |
0.1225590495 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 12 |
Hb_000731_290 |
0.1227802196 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_002811_190 |
0.1234701588 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 14 |
Hb_001405_080 |
0.127290814 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 15 |
Hb_001821_060 |
0.1280258094 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 16 |
Hb_000270_710 |
0.1313954328 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_030312_030 |
0.1314893118 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010632_010 |
0.1324708661 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000032_600 |
0.1327651869 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 20 |
Hb_001365_010 |
0.1330810109 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |