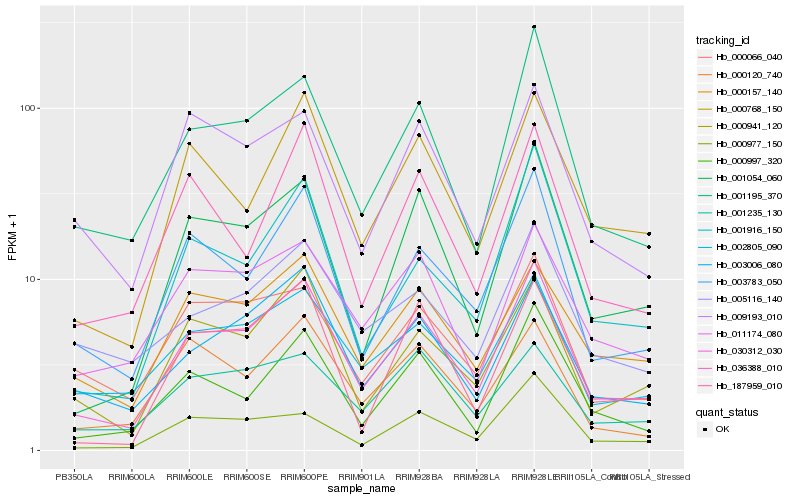
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030312_030 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001195_370 |
0.0905756527 |
- |
- |
- |
| 3 |
Hb_003783_050 |
0.098099498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001054_060 |
0.1030703832 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 5 |
Hb_000941_120 |
0.1043646494 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 6 |
Hb_003006_080 |
0.1049710086 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002805_090 |
0.1058585594 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000997_320 |
0.1108813447 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 9 |
Hb_000768_150 |
0.1117963255 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000157_140 |
0.1152682746 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_011174_080 |
0.1169524399 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 12 |
Hb_000066_040 |
0.1189893469 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 13 |
Hb_001916_150 |
0.1215082761 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 14 |
Hb_005116_140 |
0.1266225439 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 15 |
Hb_036388_010 |
0.1282868654 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 16 |
Hb_187959_010 |
0.129131806 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 17 |
Hb_000977_150 |
0.129686428 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 18 |
Hb_000120_740 |
0.1301836783 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001235_130 |
0.1306131982 |
- |
- |
- |
| 20 |
Hb_009193_010 |
0.1313971333 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |