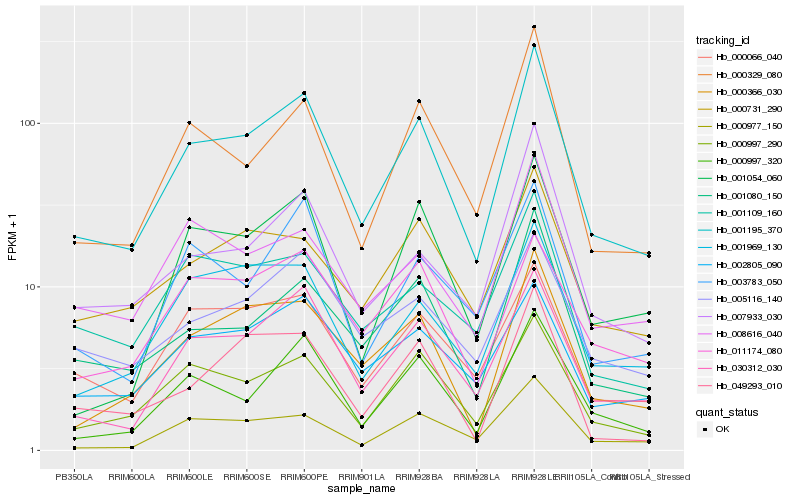
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_370 |
0.0 |
- |
- |
- |
| 2 |
Hb_030312_030 |
0.0905756527 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000997_290 |
0.0994463557 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 4 |
Hb_000329_080 |
0.1050088576 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000997_320 |
0.1065701654 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 6 |
Hb_000366_030 |
0.1067690397 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 7 |
Hb_001080_150 |
0.1068263166 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 8 |
Hb_000977_150 |
0.1161066092 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 9 |
Hb_000066_040 |
0.1190284639 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 10 |
Hb_007933_030 |
0.1227674635 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 11 |
Hb_001969_130 |
0.1232944037 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 12 |
Hb_001054_060 |
0.1253998536 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 13 |
Hb_000731_290 |
0.1257636692 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_003783_050 |
0.1284938061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008616_040 |
0.1289008885 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002805_090 |
0.1289190489 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 17 |
Hb_005116_140 |
0.1291259348 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 18 |
Hb_001109_160 |
0.1312024091 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_049293_010 |
0.1336890432 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_011174_080 |
0.1338025282 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |