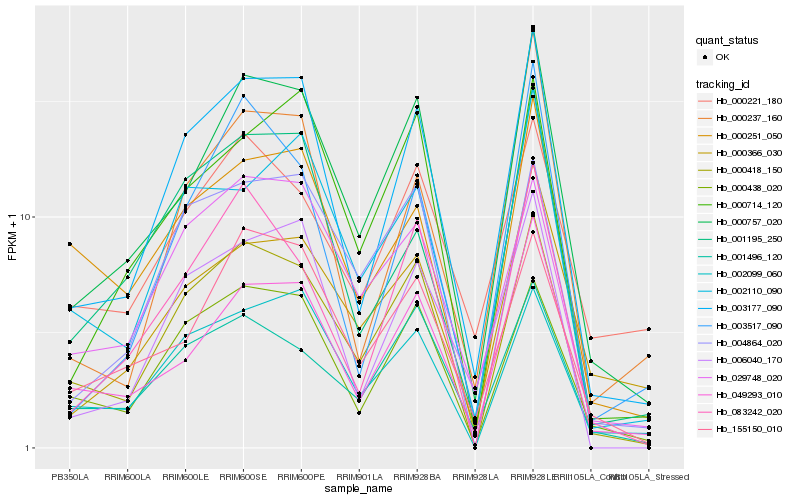
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000757_020 |
0.0 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_049293_010 |
0.0782835676 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_003177_090 |
0.1050375082 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 4 |
Hb_000418_150 |
0.1065724017 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_155150_010 |
0.1089183893 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_000714_120 |
0.1186460447 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 7 |
Hb_000366_030 |
0.1282378372 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 8 |
Hb_083242_020 |
0.1332337925 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 9 |
Hb_001496_120 |
0.1427106157 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000438_020 |
0.1454764284 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 11 |
Hb_002110_090 |
0.1519606979 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 12 |
Hb_000251_050 |
0.1522720152 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 13 |
Hb_002099_060 |
0.1524609124 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 14 |
Hb_029748_020 |
0.1536808408 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_004864_020 |
0.1543178878 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 16 |
Hb_003517_090 |
0.1567154435 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_006040_170 |
0.1574667513 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 18 |
Hb_001195_250 |
0.1597695672 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000221_180 |
0.161940122 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 20 |
Hb_000237_160 |
0.1622315295 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |