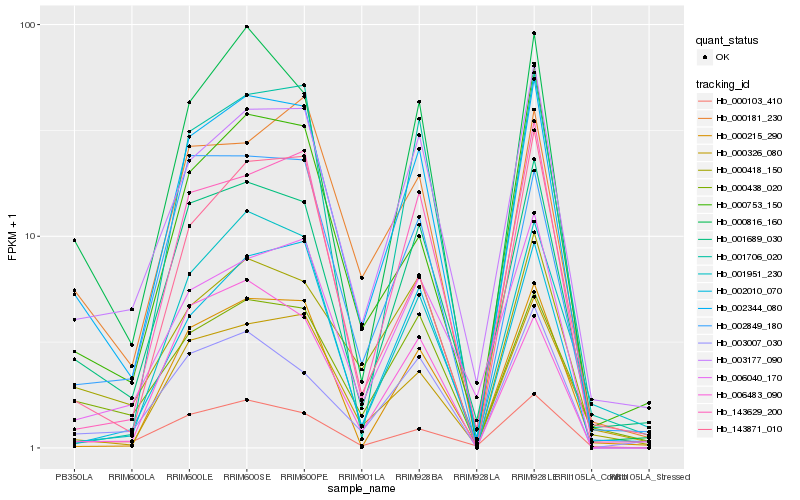
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002344_080 |
0.0 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 2 |
Hb_001689_030 |
0.0793072091 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 3 |
Hb_000816_160 |
0.1008391168 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 4 |
Hb_003177_090 |
0.1028272352 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 5 |
Hb_000181_230 |
0.1138599056 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_143629_200 |
0.1146587031 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 7 |
Hb_000103_410 |
0.1212119613 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 8 |
Hb_002010_070 |
0.1237481694 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 9 |
Hb_000418_150 |
0.1255567253 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_002849_180 |
0.1256484668 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 11 |
Hb_000326_080 |
0.1257316117 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000438_020 |
0.1282496787 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 13 |
Hb_006040_170 |
0.1291788046 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 14 |
Hb_000215_290 |
0.1324824466 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003007_030 |
0.1332884575 |
- |
- |
- |
| 16 |
Hb_143871_010 |
0.1368450483 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_001706_020 |
0.141530145 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 18 |
Hb_006483_090 |
0.1428259834 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_000753_150 |
0.1431784492 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 20 |
Hb_001951_230 |
0.1432003983 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |