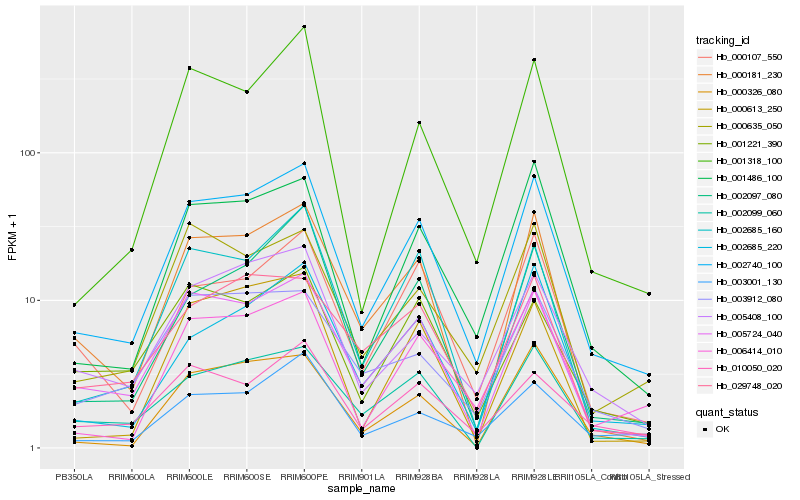
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002740_100 |
0.0 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 2 |
Hb_005724_040 |
0.0883312588 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 3 |
Hb_001486_100 |
0.0924543898 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 4 |
Hb_000181_230 |
0.1063231899 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_006414_010 |
0.1073655836 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 6 |
Hb_005408_100 |
0.1099109034 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 7 |
Hb_000326_080 |
0.1171012098 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000107_550 |
0.1171267609 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 9 |
Hb_002685_160 |
0.1181406665 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 10 |
Hb_002099_060 |
0.1187041373 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 11 |
Hb_002685_220 |
0.1210526866 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000635_050 |
0.1242345591 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 13 |
Hb_003001_130 |
0.1265405219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001318_100 |
0.1284736833 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 15 |
Hb_002097_080 |
0.1285226417 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 16 |
Hb_003912_080 |
0.1332640918 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 17 |
Hb_029748_020 |
0.1333991293 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001221_390 |
0.134230703 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 19 |
Hb_000613_250 |
0.134387612 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 20 |
Hb_010050_020 |
0.1344555612 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |