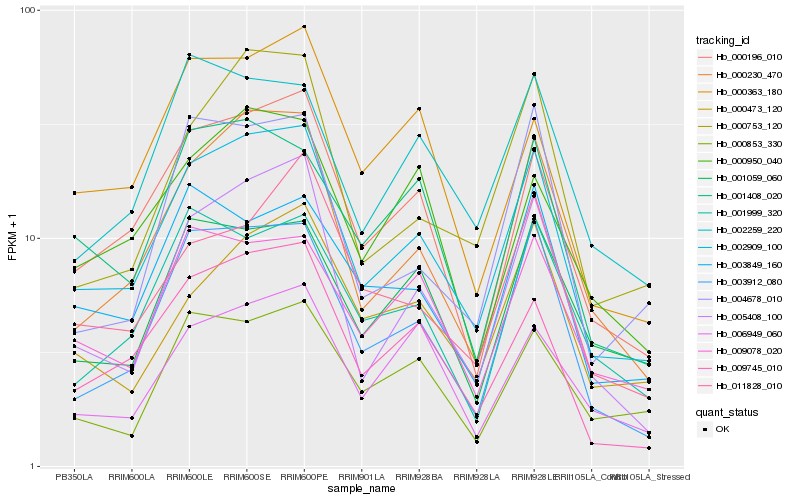
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002909_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 2 |
Hb_000230_470 |
0.0844250237 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000196_010 |
0.0875529507 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 4 |
Hb_003912_080 |
0.0886062837 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 5 |
Hb_009745_010 |
0.0936738615 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 6 |
Hb_005408_100 |
0.0977469555 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 7 |
Hb_000753_120 |
0.1080138512 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 8 |
Hb_009078_020 |
0.1168370115 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 9 |
Hb_001408_020 |
0.1189167324 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 10 |
Hb_002259_220 |
0.1220088463 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 11 |
Hb_001059_060 |
0.1220298087 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 12 |
Hb_003849_160 |
0.1220527696 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 13 |
Hb_000473_120 |
0.1225240401 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 14 |
Hb_001999_320 |
0.1243802966 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 15 |
Hb_004678_010 |
0.1254823608 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 16 |
Hb_000853_330 |
0.1254901224 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 17 |
Hb_011828_010 |
0.1261147836 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 18 |
Hb_000363_180 |
0.1266171844 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 19 |
Hb_006949_060 |
0.1268323974 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 20 |
Hb_000950_040 |
0.1274019458 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |