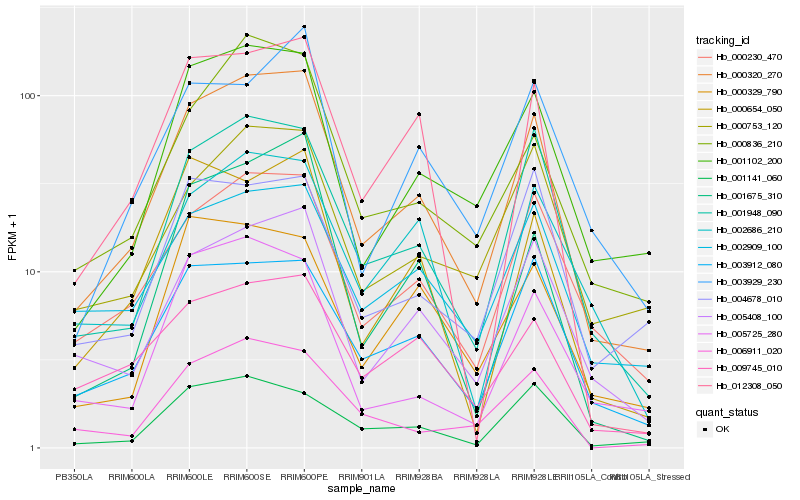
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_270 |
0.0 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001948_090 |
0.0982374326 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 3 |
Hb_005408_100 |
0.1077592873 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 4 |
Hb_000230_470 |
0.1079890896 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001102_200 |
0.1079957324 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 6 |
Hb_003912_080 |
0.1229548105 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_012308_050 |
0.1285476452 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 8 |
Hb_000836_210 |
0.1295116223 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 9 |
Hb_000654_050 |
0.1308489926 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 10 |
Hb_001675_310 |
0.1328266905 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001141_060 |
0.1334775204 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000753_120 |
0.1337033113 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 13 |
Hb_002909_100 |
0.1343402445 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 14 |
Hb_009745_010 |
0.1370609493 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 15 |
Hb_002686_210 |
0.1389889658 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_005725_280 |
0.1390429416 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 17 |
Hb_006911_020 |
0.1409881983 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_003929_230 |
0.1433013418 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 19 |
Hb_000329_790 |
0.1448636036 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 20 |
Hb_004678_010 |
0.1488135771 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |