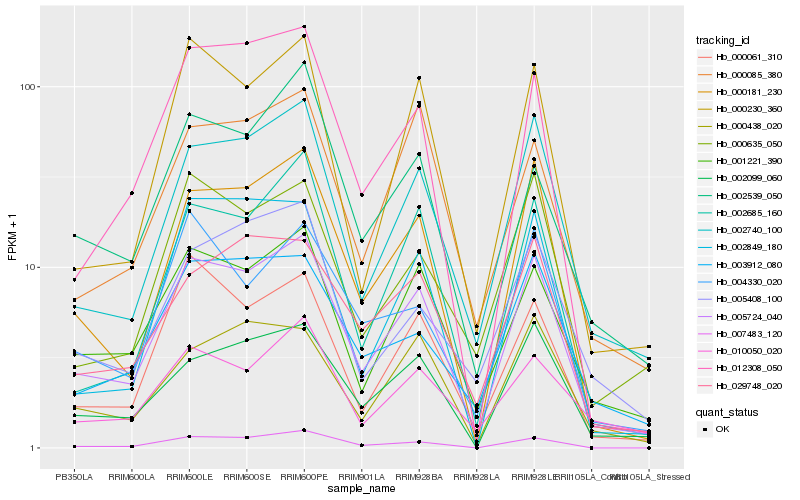
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005724_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 2 |
Hb_000181_230 |
0.0873595282 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_002740_100 |
0.0883312588 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_001221_390 |
0.0970087309 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 5 |
Hb_007483_120 |
0.1018295238 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 6 |
Hb_002099_060 |
0.105763496 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_002685_160 |
0.1145275027 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000230_360 |
0.1151500023 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 9 |
Hb_000061_310 |
0.1202455424 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_029748_020 |
0.1260919142 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_012308_050 |
0.1265562197 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 12 |
Hb_000635_050 |
0.1289007908 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 13 |
Hb_002849_180 |
0.1304739705 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 14 |
Hb_000438_020 |
0.1307585548 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 15 |
Hb_003912_080 |
0.131038382 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 16 |
Hb_010050_020 |
0.1313544799 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 17 |
Hb_000085_380 |
0.1348856276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004330_020 |
0.1400169868 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 19 |
Hb_005408_100 |
0.1415798201 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 20 |
Hb_002539_050 |
0.1421212399 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |