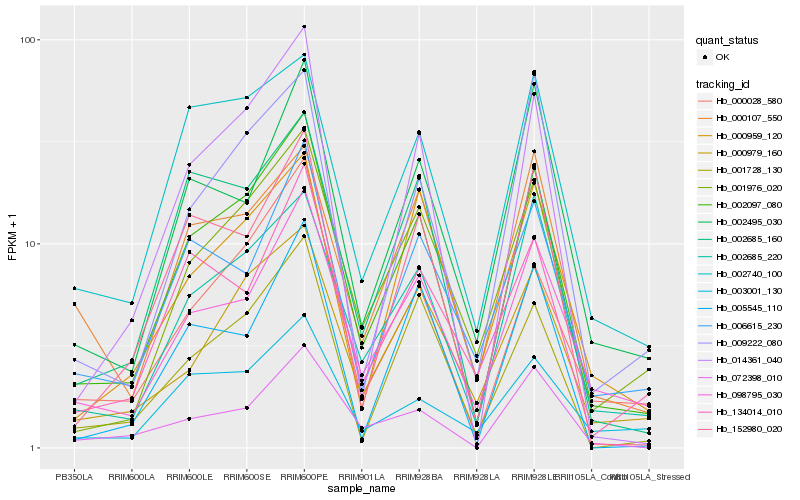
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002097_080 |
0.0 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 2 |
Hb_002685_220 |
0.1080066131 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001976_020 |
0.110361031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_014361_040 |
0.1117665575 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 5 |
Hb_098795_030 |
0.113027725 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 6 |
Hb_002685_160 |
0.1180634117 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000028_580 |
0.1188488807 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 8 |
Hb_001728_130 |
0.1257094636 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_152980_020 |
0.1270828171 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 10 |
Hb_002740_100 |
0.1285226417 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 11 |
Hb_009222_080 |
0.1327302556 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 12 |
Hb_000979_160 |
0.1334790297 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 13 |
Hb_002495_030 |
0.1367520845 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006615_230 |
0.1385546966 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 15 |
Hb_072398_010 |
0.1386649114 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 16 |
Hb_000959_120 |
0.1427300798 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 17 |
Hb_134014_010 |
0.1453809321 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_005545_110 |
0.1467848212 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 19 |
Hb_003001_130 |
0.1480251215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000107_550 |
0.1487830204 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |