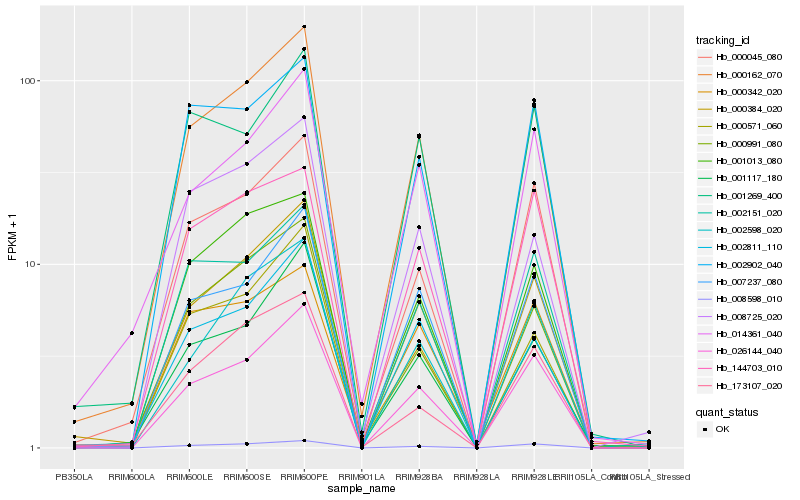
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_070 |
0.0 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 2 |
Hb_026144_040 |
0.0672082393 |
- |
- |
- |
| 3 |
Hb_008598_010 |
0.0798723356 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 4 |
Hb_007237_080 |
0.08369774 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_000045_080 |
0.0847898353 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_000991_080 |
0.085352426 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_002811_110 |
0.0857562988 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 8 |
Hb_000384_020 |
0.0860921044 |
- |
- |
sucrose phosphate syntase, putative [Ricinus communis] |
| 9 |
Hb_001013_080 |
0.0870020012 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 10 |
Hb_002598_020 |
0.092983808 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 [Populus euphratica] |
| 11 |
Hb_001117_180 |
0.0937709907 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 12 |
Hb_014361_040 |
0.0962444567 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 13 |
Hb_002151_020 |
0.0988232793 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 14 |
Hb_008725_020 |
0.1026107386 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 15 |
Hb_000571_060 |
0.1099420758 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 16 |
Hb_173107_020 |
0.1100833173 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 17 |
Hb_000342_020 |
0.1105448138 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 18 |
Hb_002902_040 |
0.1115234165 |
- |
- |
PREDICTED: peroxidase 51 [Jatropha curcas] |
| 19 |
Hb_144703_010 |
0.1126043466 |
- |
- |
Phospholipase A 2A, IIA,PLA2A [Theobroma cacao] |
| 20 |
Hb_001269_400 |
0.1127907381 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |