| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
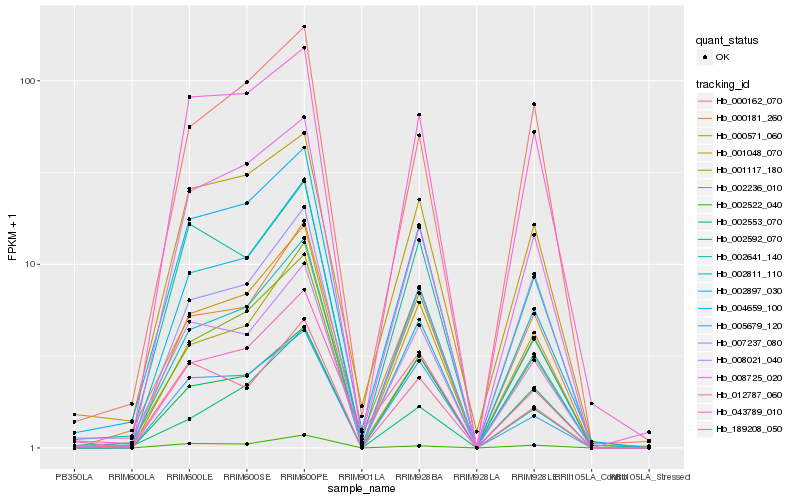
Hb_000571_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 2 |
Hb_002811_110 |
0.0514907411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 3 |
Hb_002236_010 |
0.0914922077 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 4 |
Hb_002553_070 |
0.096170422 |
- |
- |
PREDICTED: RING-H2 finger protein ATL16 [Jatropha curcas] |
| 5 |
Hb_000181_260 |
0.0967094303 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 6 |
Hb_007237_080 |
0.0979508873 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_008725_020 |
0.1002249363 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 8 |
Hb_012787_060 |
0.1071781583 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 9 |
Hb_008021_040 |
0.1079080694 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 10 |
Hb_002897_030 |
0.1081688178 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 11 |
Hb_043789_010 |
0.1085931157 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 12 |
Hb_000162_070 |
0.1099420758 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 13 |
Hb_005679_120 |
0.1105546409 |
- |
- |
hypothetical protein RCOM_0284080 [Ricinus communis] |
| 14 |
Hb_001117_180 |
0.1116636338 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002522_040 |
0.1162772277 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 16 |
Hb_004659_100 |
0.1171473863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_189208_050 |
0.1172102394 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001048_070 |
0.1207846757 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 19 |
Hb_002592_070 |
0.1229279738 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 20 |
Hb_002641_140 |
0.1237783546 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |