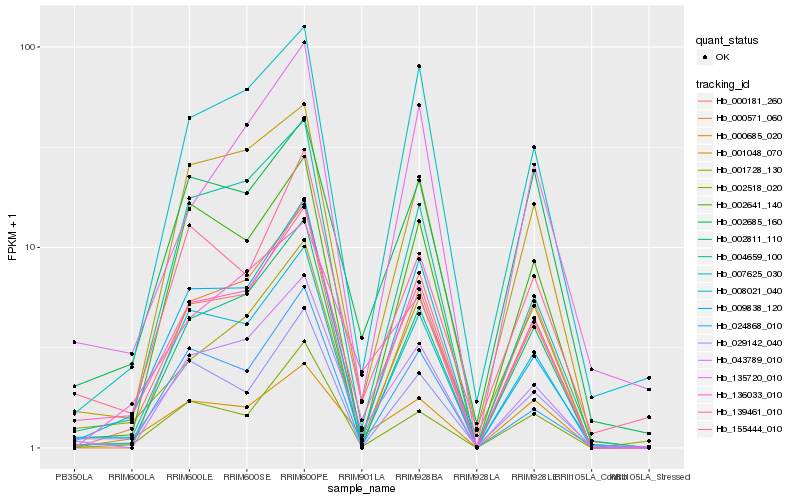
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139461_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 2 |
Hb_008021_040 |
0.0847100934 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 3 |
Hb_000181_260 |
0.0919176519 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 4 |
Hb_043789_010 |
0.0980334531 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 5 |
Hb_002811_110 |
0.1093777308 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 6 |
Hb_009838_120 |
0.1125495075 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001048_070 |
0.1238830877 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 8 |
Hb_029142_040 |
0.1268902044 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 9 |
Hb_007625_030 |
0.1293654958 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_155444_010 |
0.1305554525 |
- |
- |
- |
| 11 |
Hb_000685_020 |
0.1308891827 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 12 |
Hb_135720_010 |
0.1322334748 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 13 |
Hb_002518_020 |
0.133273309 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 14 |
Hb_000571_060 |
0.1356860743 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 15 |
Hb_024868_010 |
0.135817033 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_136033_010 |
0.1360559115 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Gossypium raimondii] |
| 17 |
Hb_001728_130 |
0.1373794409 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_004659_100 |
0.1384752411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002685_160 |
0.1393055671 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 20 |
Hb_002641_140 |
0.1400022979 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |