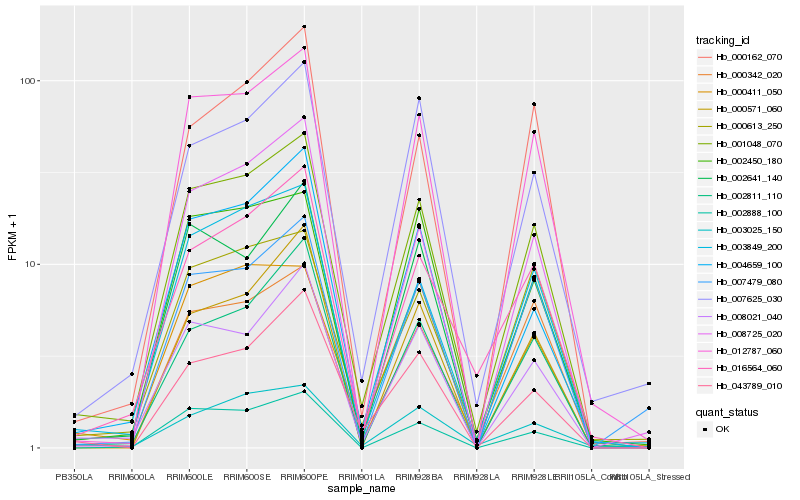
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001048_070 |
0.0 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 2 |
Hb_012787_060 |
0.0764503305 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 3 |
Hb_003849_200 |
0.0852488647 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_008021_040 |
0.0933828047 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 5 |
Hb_002888_100 |
0.0989531898 |
- |
- |
PREDICTED: uncharacterized protein LOC103699074, partial [Phoenix dactylifera] |
| 6 |
Hb_002641_140 |
0.0997278481 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 7 |
Hb_000342_020 |
0.1039581084 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 8 |
Hb_016564_060 |
0.1048555796 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 9 |
Hb_008725_020 |
0.1083180386 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 10 |
Hb_000613_250 |
0.1094673025 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 11 |
Hb_002811_110 |
0.1102685012 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 12 |
Hb_002450_180 |
0.1105289551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004659_100 |
0.110616573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007625_030 |
0.1147405105 |
- |
- |
unknown [Medicago truncatula] |
| 15 |
Hb_007479_080 |
0.1147707061 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 16 |
Hb_043789_010 |
0.1151994622 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 17 |
Hb_003025_150 |
0.1177949298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000411_050 |
0.118049007 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 19 |
Hb_000162_070 |
0.1206480903 |
- |
- |
phospholipase C, putative [Ricinus communis] |
| 20 |
Hb_000571_060 |
0.1207846757 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |