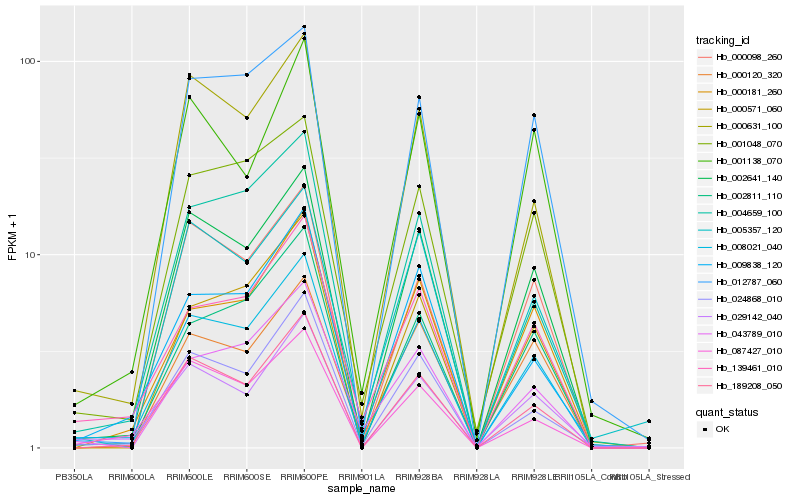
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008021_040 |
0.0 |
- |
- |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 2 |
Hb_002641_140 |
0.0664698396 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_139461_010 |
0.0847100934 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 4 |
Hb_189208_050 |
0.085494839 |
- |
- |
PREDICTED: uncharacterized protein LOC105642694 isoform X2 [Jatropha curcas] |
| 5 |
Hb_024868_010 |
0.0890131156 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_001048_070 |
0.0933828047 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 7 |
Hb_029142_040 |
0.0946121279 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 8 |
Hb_002811_110 |
0.0950194678 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 9 |
Hb_000631_100 |
0.0967205138 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 10 |
Hb_009838_120 |
0.0987761344 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000181_260 |
0.0993758719 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 12 |
Hb_004659_100 |
0.1033978947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_043789_010 |
0.1057445489 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 14 |
Hb_000571_060 |
0.1079080694 |
- |
- |
PREDICTED: uncharacterized protein LOC105648112 [Jatropha curcas] |
| 15 |
Hb_087427_010 |
0.1084005822 |
- |
- |
hypothetical protein PRUPE_ppa007748mg [Prunus persica] |
| 16 |
Hb_001138_070 |
0.115030117 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_012787_060 |
0.1157373701 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |
| 18 |
Hb_005357_120 |
0.1167699974 |
transcription factor |
TF Family: MYB-related |
- |
| 19 |
Hb_000098_260 |
0.1170832004 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 20 |
Hb_000120_320 |
0.1172427029 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |